-
Notifications
You must be signed in to change notification settings - Fork 2
Learning about the KB
-
Balhoff JP, Phenoscape Project Team. The Phenoscape Knowledgebase: tools and APIs for computing across phenotypes from evolutionary diversity and model organisms. bioRxiv. 2016. p. 071951. doi:10.1101/071951
A short read (2 pages) that gives an overview of the data sources and ontologies going into building the KB, the tools and steps involved in building, and the web-service interfaces.
-
Dececchi TA, Balhoff JP, Lapp H, Mabee PM. Toward Synthesizing Our Knowledge of Morphology: Using Ontologies and Machine Reasoning to Extract Presence/Absence Evolutionary Phenotypes across Studies. Syst Biol. 2015;64: 936–952. doi:10.1093/sysbio/syv031
Paper describing how the KB uses machine reasoning to synthesize presence/absence characters that are implied but not expressly stated by published phenotype descriptions.
-
Manda P, Balhoff JP, Lapp H, Mabee P, Vision TJ. Using the phenoscape knowledgebase to relate genetic perturbations to phenotypic evolution. Genesis. 2015;53: 561–571. doi:10.1002/dvg.22878
Paper describing how algorithms for calculating semantic similarity and significance allow obtaining taxa with evolutionary phenotype transitions semantically similar to the phenotypes of a gene when mutated or knocked out.
-
Most current and complete documentation of the KB web-service API: http://kb.phenoscape.org/apidocs/
Note that this API is also what the Phenoscape KB online UI uses for data access. Therefore, data that appears on the web UI but for which there appears to be no documented API method is likely from an API method not yet documented.
-
Examples for API clients other than the Phenoscape KB UI:
- RPhenoscape (see also talk from 2016 Evolution Meetings)
-
Most API methods return data in JSON format. The structure of the returned data for each of the services is documented in the REST API documentation (see above). Typically, the identifier for a data item is returned as an IRI in the value of the
@idproperty. Services that return lists of results place the list inside a top-levelresultsproperty. Services that support paging of results will return the total items available (instead of returning results) when thetotal=trueparameter is included. These results will return a single integer as the value of thetotalproperty.Several services support returning TSV via content-negotiation (documented in the response section of the API documentation). TSV can be obtained by requesting the
text/plaincontent type. If JSON is desired, anapplication/jsoncontent type should always be requested in the Accept header, since additional return formats may be added in the future. -
API methods that return a data matrix return data in NeXML format. The following papers may be useful in this regard:
-
Vos RA, Balhoff JP, Caravas JA, Holder MT, Lapp H, Maddison WP, et al. NeXML: rich, extensible, and verifiable representation of comparative data and metadata. Syst Biol. 2012;61: 675–689. doi:10.1093/sysbio/sys025
A description of the NeXML format as an exchange standard for comparative evolutionary analysis.
-
Boettiger C, Chamberlain S, Vos R, Lapp H. RNeXML: a package for reading and writing richly annotated phylogenetic, character and trait data in R. Methods Ecol Evol. 2016;7: 352–357. doi:10.1111/2041-210X.12469
A description of how the R package RNeXML does the heavy lifting of parsing NeXML in R to make it easy to consume for R users. Unfortunately there currently aren't comparable packages/libraries in other languages.
-
-
The Phenoscape KB UI includes an experimental data content visualization interface that allows interactively exploring how the data content distributes along certain axes (right now these axes are taxonomy and anatomy). Examples as static images (top: data content across groups within otophysi; bottom: data content across parts of the neurocranium):
-
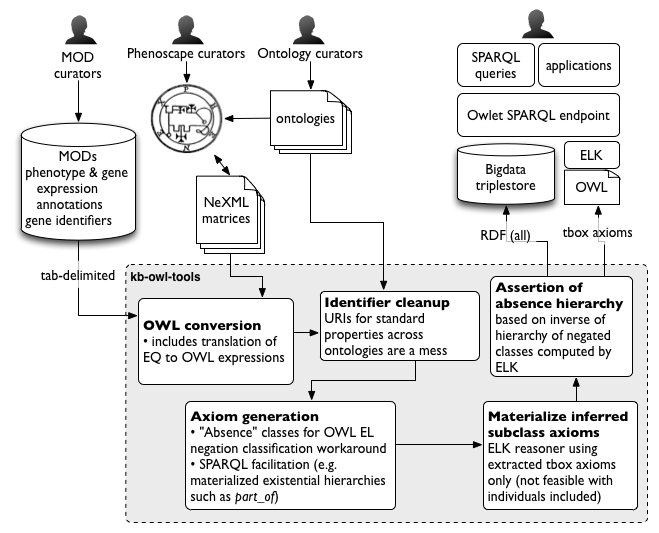
Schematic diagram of the data flow into and out of the KB (Figure 1 in Balhoff et al, 2016), which is from the Phenoscape Wiki page on the KB build process:
-
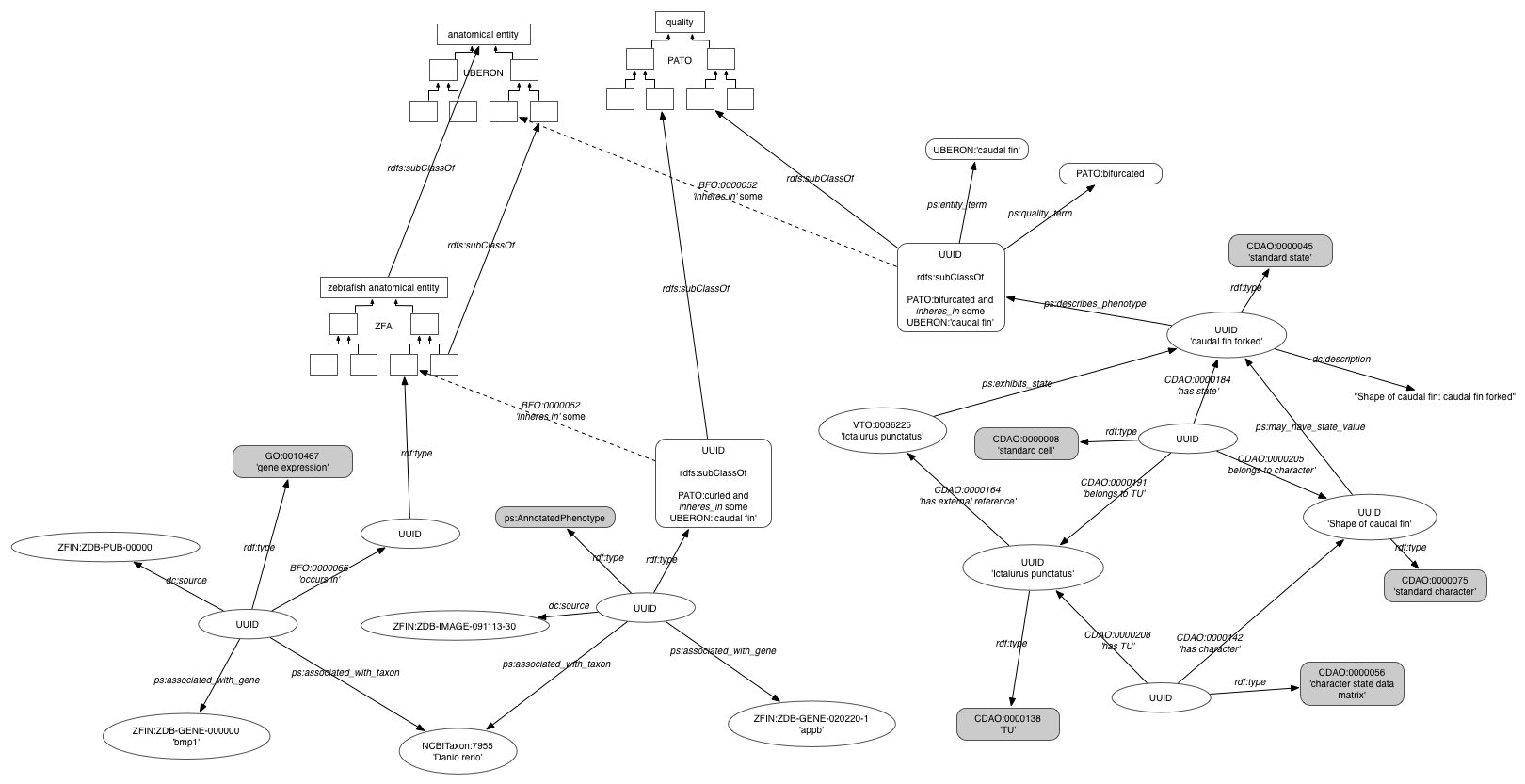
Schematic of the triple model in which data are represented in the KB (high-res version):
Sponsored by Phenoscape, a project funded by NSF grants DBI-1062404 and DBI-1062542. Phenoscape was incubated and supported by the National Evolutionary Synthesis Center (NESCent), NSF EF-0905606.