- + hemibrain and flywire meta data + +
- flywire
-
@@ -108,9 +111,6 @@
flywire
--
“We’re Building Something, Here, Detective, We’re Building It From Scratch. All The Pieces Matter.” (Lester, The Wire)
-FlyWire and R
@@ -122,7 +122,7 @@- Read flywire NBLASTs and NBLASTs to hemibrain neurons
- Read flywire neurons that are pre-transformed into a variety of brainspaces
-Which is all useful stuff. In order to connect R to this Google drive, you have a few options. Please see this article. The pipeline that produced this data can be found here. It is run nightly on a machine at the MRC LMB.
+Which is all useful stuff. In order to connect R to this Google drive, you have a few options. Please see this article. The pipeline that produced this data can be found here. It is run nightly on a machine at the MRC LMB.
-Authorisation
@@ -162,9 +162,9 @@if (!require("fafbseg")) remotes::install_github("flyconnectome/fafbseg") library(nat.jrcbrains) library(fafbseg)
+diff --git a/docs/articles/google_filestream.html b/docs/articles/google_filestream.html index 55e916dd..bfb0edef 100644 --- a/docs/articles/google_filestream.html +++ b/docs/articles/google_filestream.html @@ -55,6 +55,9 @@-Get flywire
+Get flywire dataWith
hemibrainryou can easily get thousands of skeletons for flywire neurons. These skeletons have been built using skeletor, which is a python module. You can also use it directly in R withfafbseg::skeletor. It has been run on thousands of neurons, which have then been stored on Google Drive, at:hemibrainr/flywire_neurons/asnat::neuronlistfhobjects. This means that you can use either get flywire skeletons for neurons using the drive, or by making them from meshes yourself. Let’s demonstrate:@@ -172,7 +172,7 @@
From the the hemibrainr Google drive we can see which flywire IDs have been read from FlyWire and skeletonised. We can also see some meta data about them, and get a data frame that tells us which users have built these neurons - useful for assigning credit!
+head(fw.edits) + +# For flywire IDs which do not have available skeletons on the google drive: +## but have been flaghged for processing (i.e. they errored) +fw.failed = flywire_failed() +head(fw.failed)# All flywire IDs for neurons that have a split precomputed -fw.ids = flywire_ids() +fw.ids = flywire_ids(sql=FALSE) length(fw.ids) # For these flywire IDs, their meta data: @@ -181,7 +181,12 @@# For flywire IDs, which users contributed what: fw.edits = flywire_contributions() -head(fw.edits)
Now, excitingly we can read the neurons themselves! And not only that, we can read them mirrored (flipped to the other hemisphere of the brain) or in a different brainspace (from a small selection) if we wish. We read these neurons as
nat::neuronlistfhobjects. This means that a data frame for the neurons and information specifying where to find each neuron’s data is read into R - but not the whole, hugenat::neuronlistobject. This saves on memory for your R session. When an operation that requires the actual neuron data is performed, neurons are read into R.+# Get all aflywire neurons @@ -281,7 +286,7 @@Match flywire neurons to each other and the hemibrain
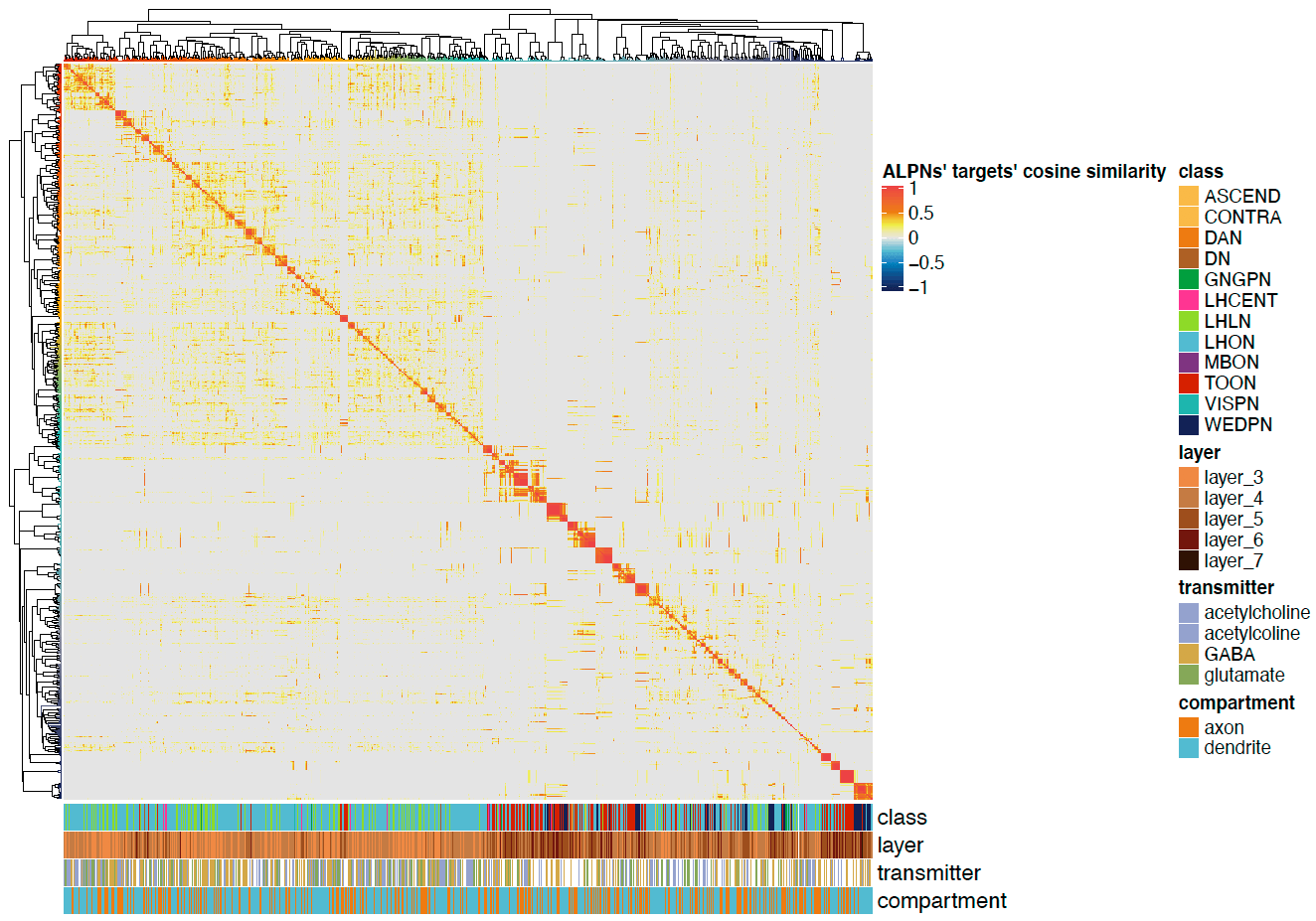
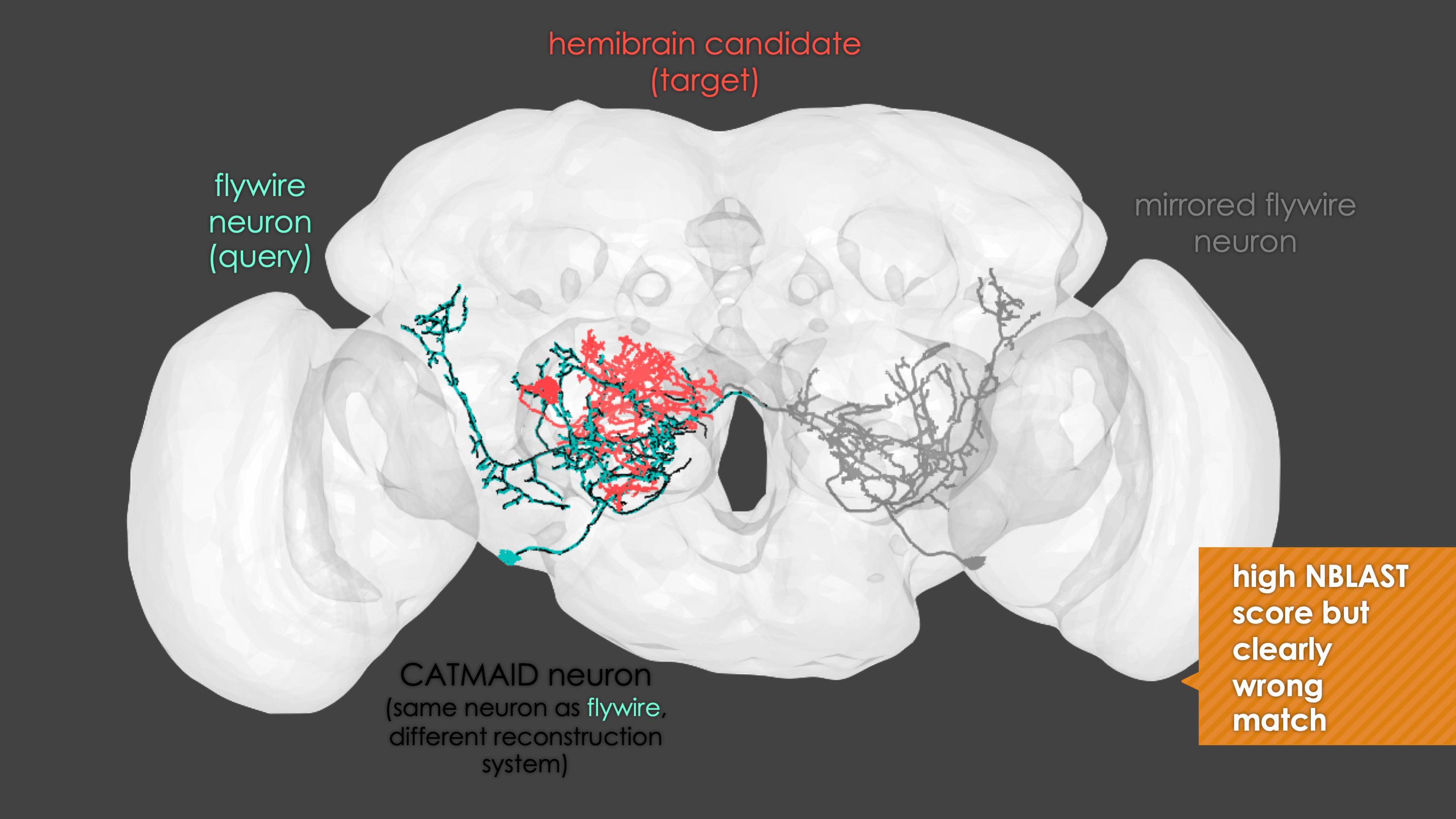
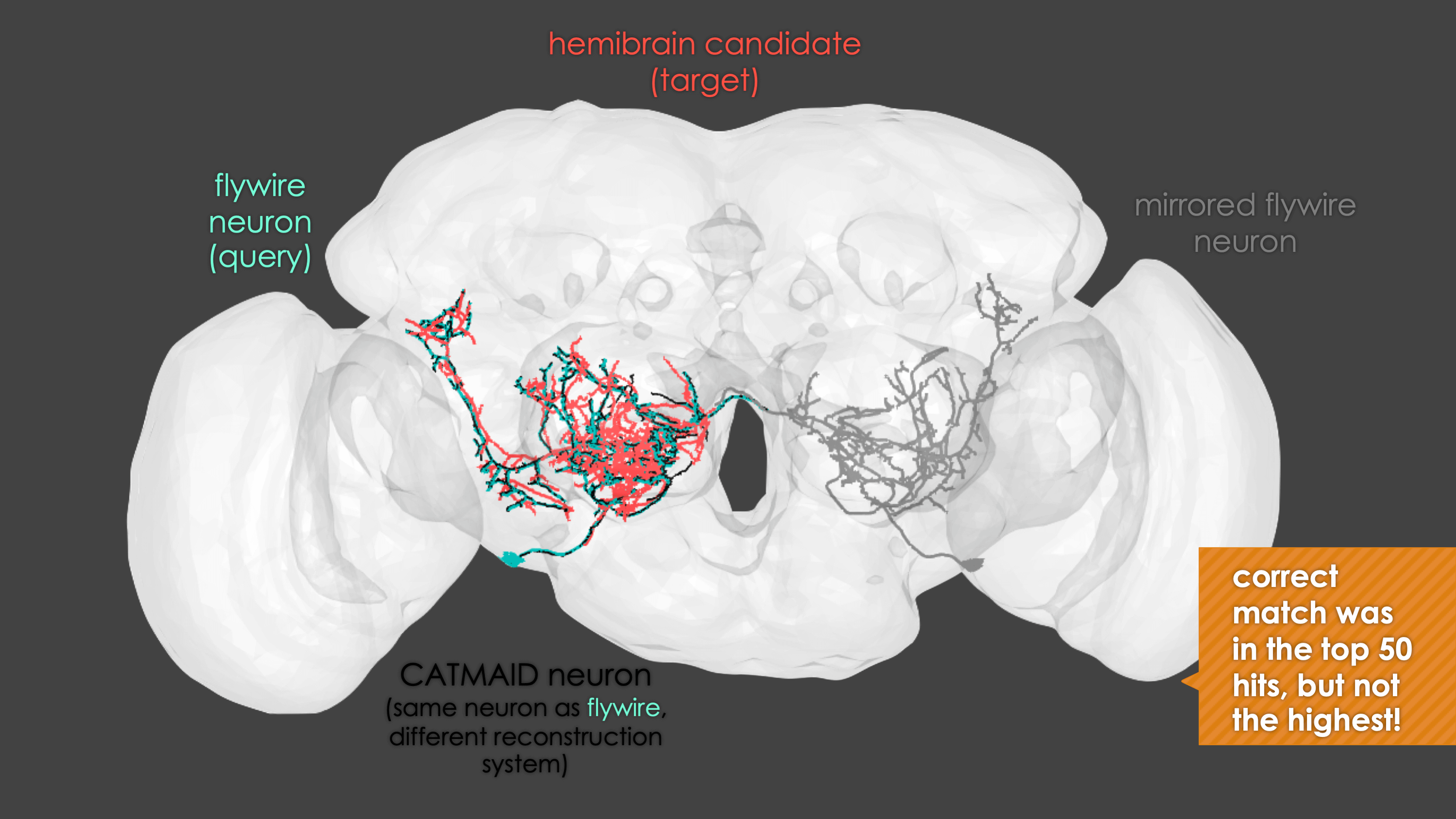
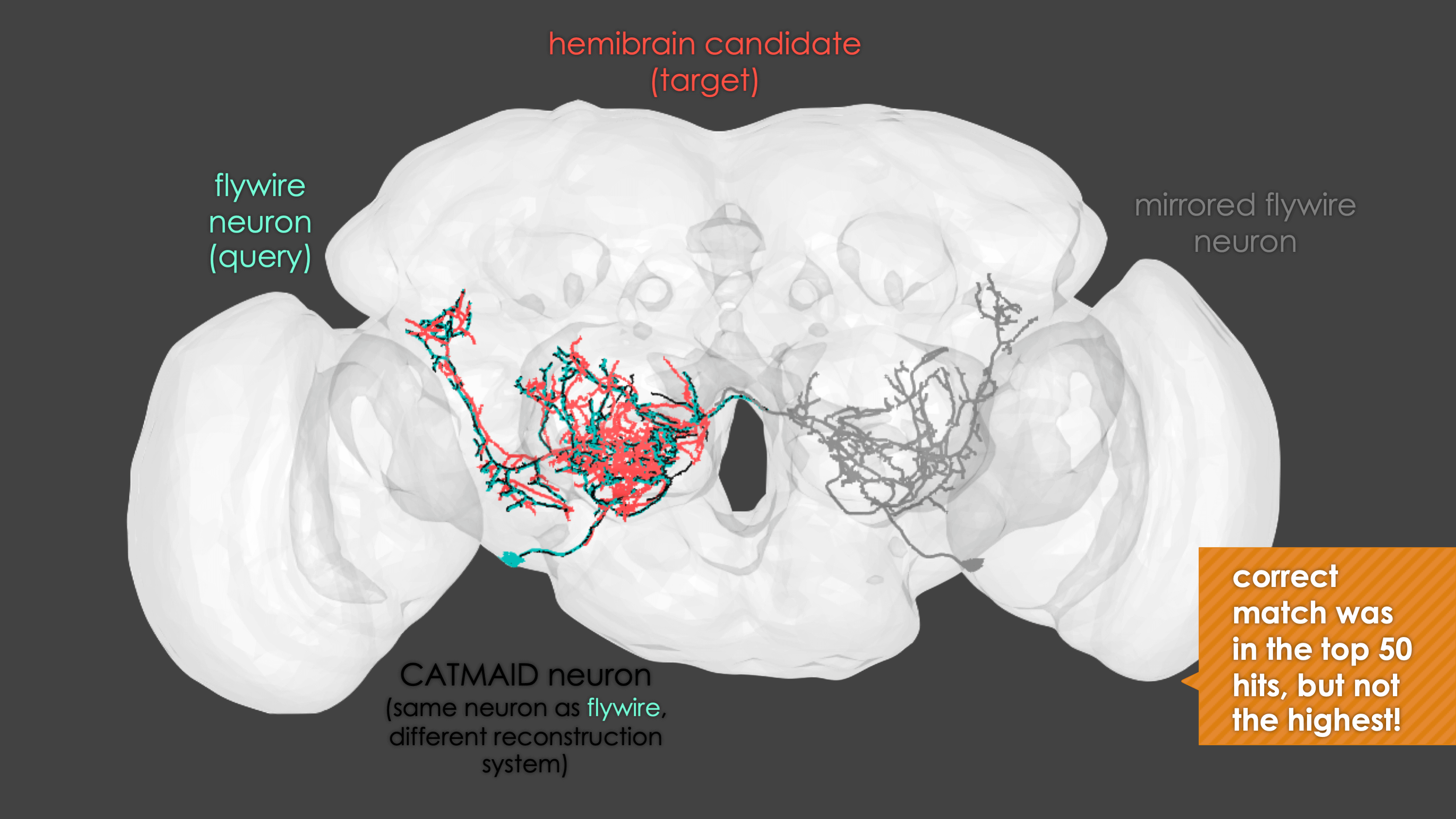
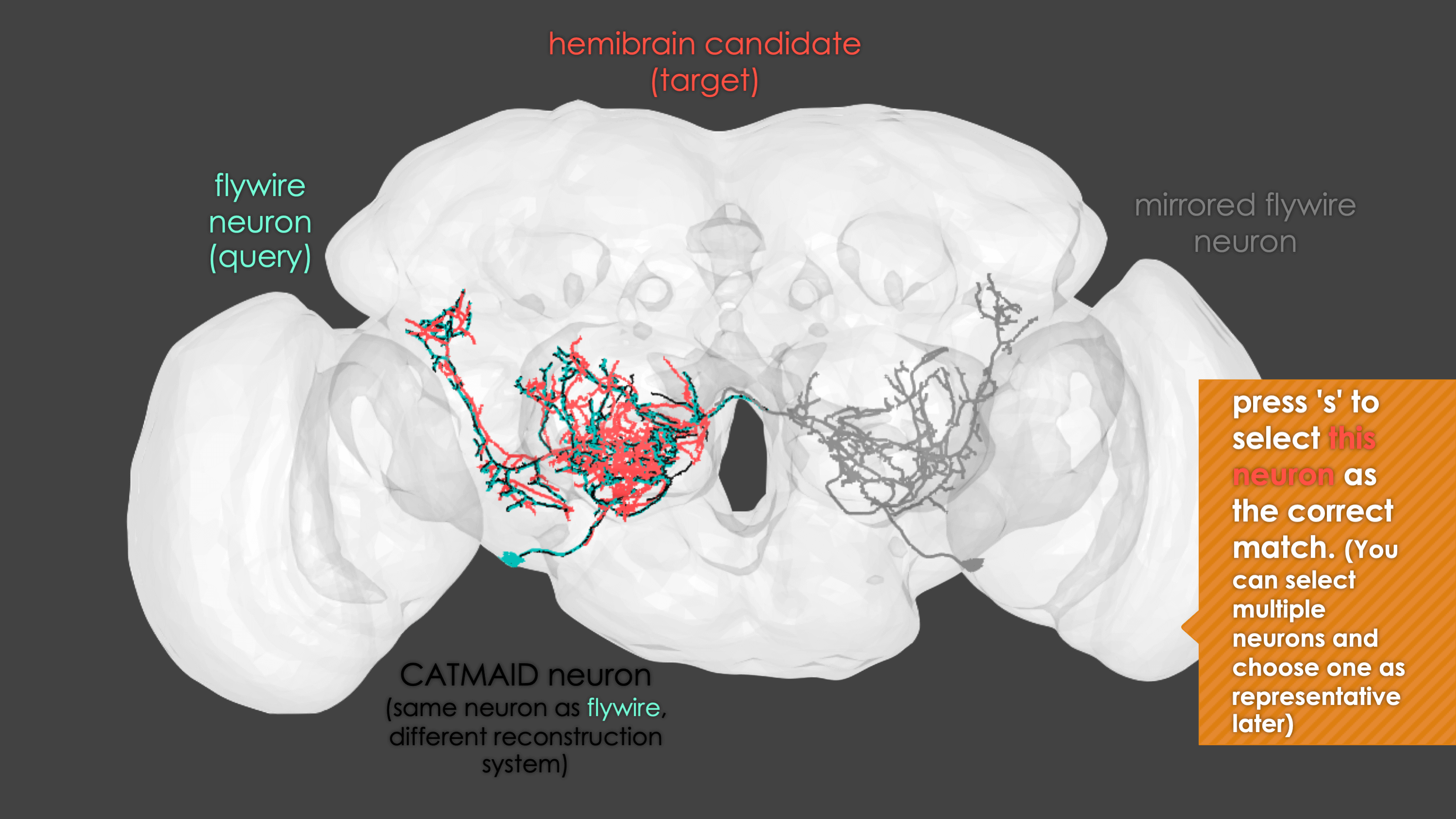
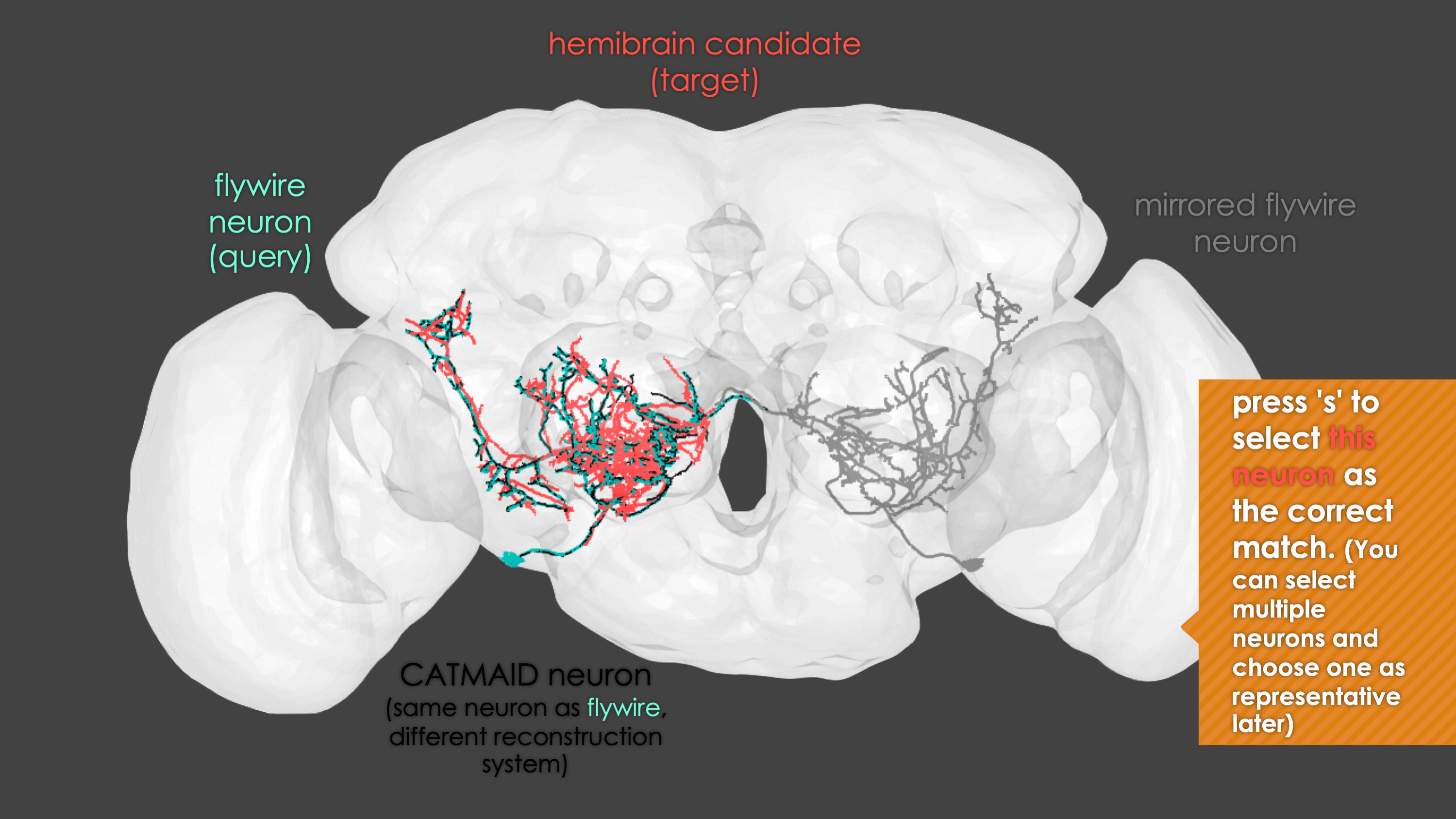
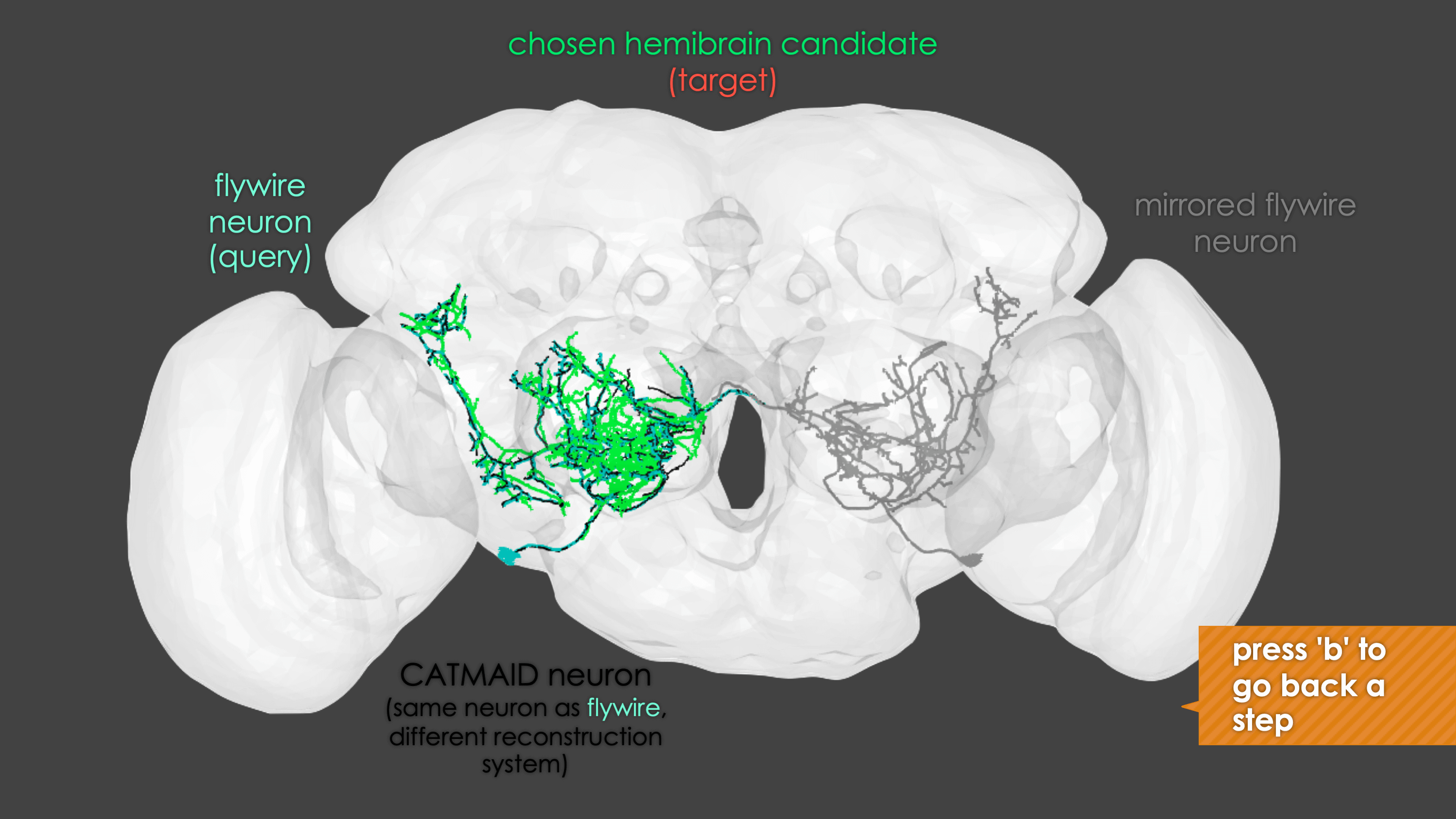
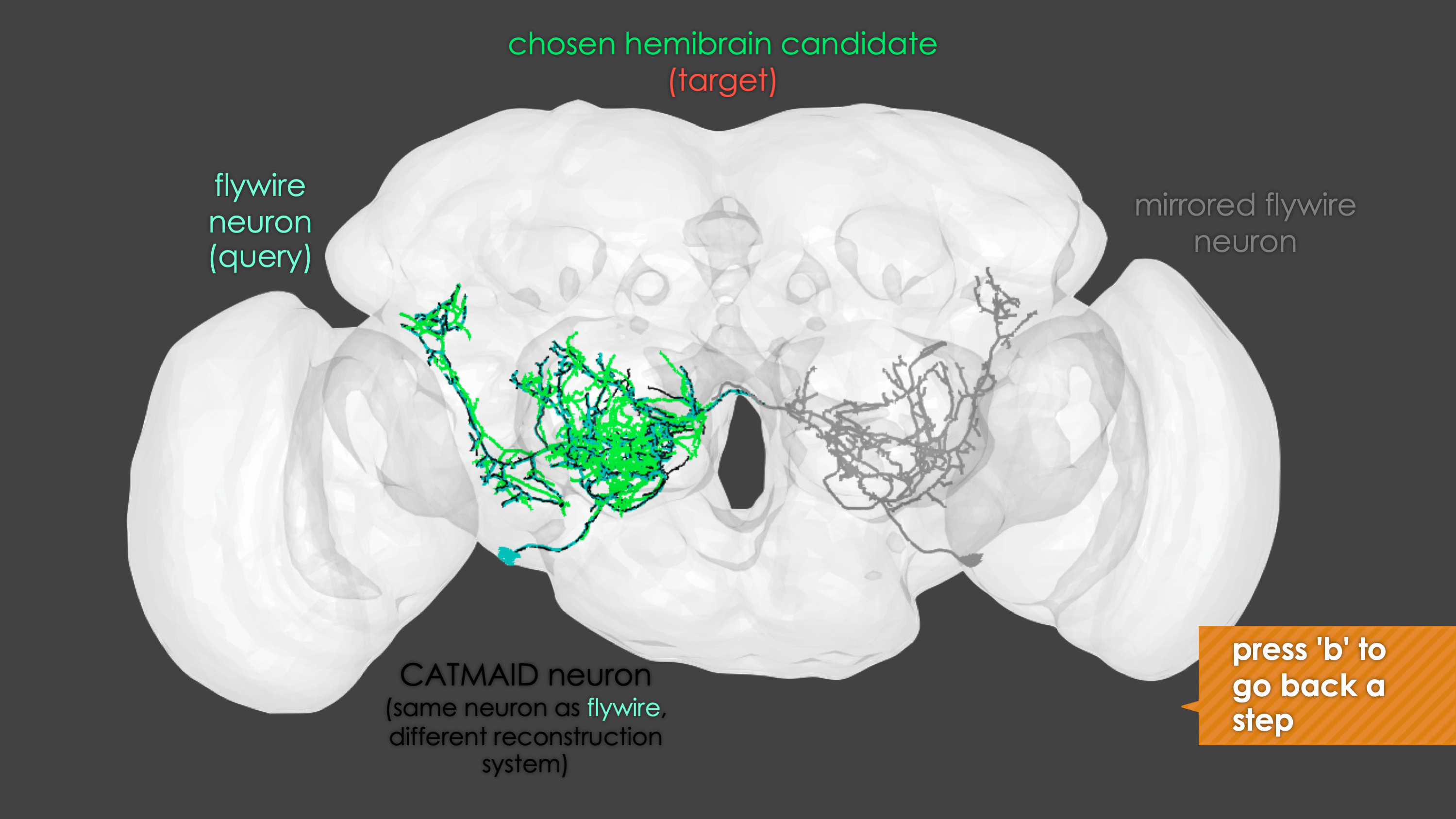
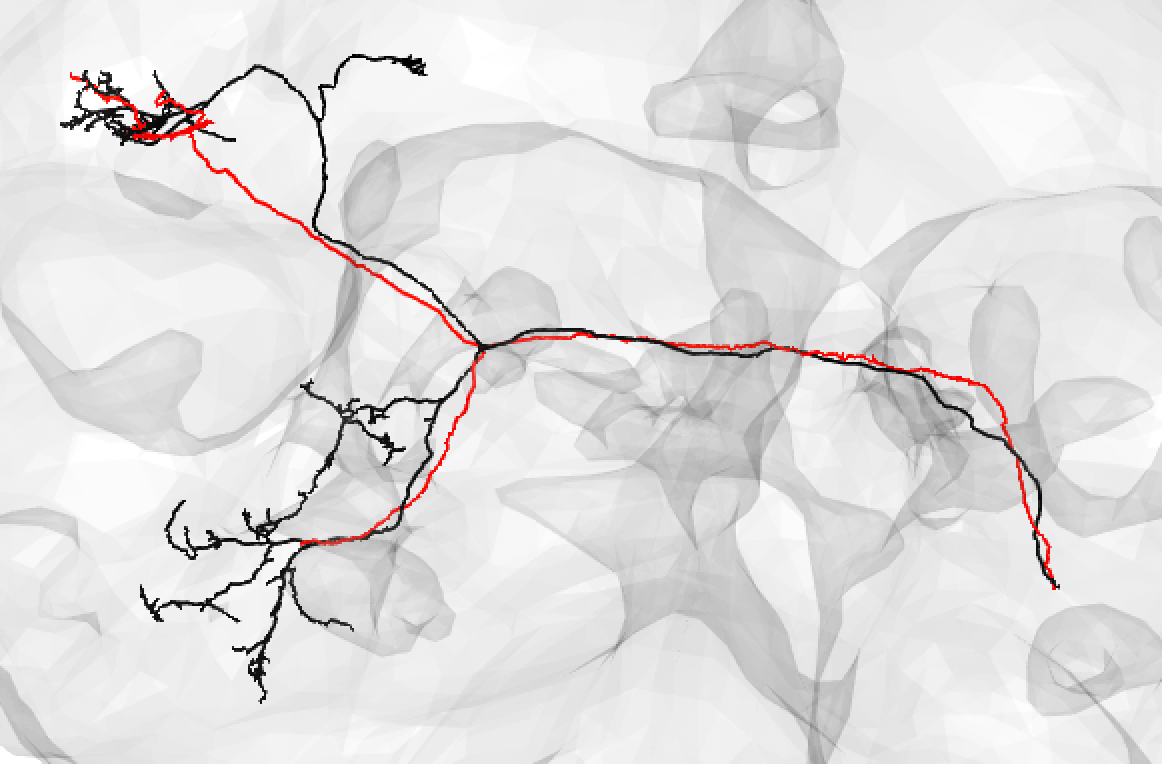
-The neuron matching pipeline reads neuron skeletons from Google Drive and and displays them. FlyWire skeletons are shown in dark grey (their mirrored equivalents in light grey) and potential hemibrain matches in red. This depends also on the precomputed NBLASTs on the hemibrain team drive. For full details, see our
+match_makingvignette.The neuron matching pipeline reads neuron skeletons from Google Drive and and displays them. FlyWire skeletons are shown in dark grey (their mirrored equivalents in light grey) and potential hemibrain matches in red. This depends also on the precomputed NBLASTs on the hemibrain team drive. For full details, see our
match_makingvignette.Matching pipelines
@@ -309,6 +314,26 @@flywire_matching_rewrite() # bring all the flywire information up to date++Full Example
+Let us get some neurons from the ‘flywire interest’ googlesheet:
+++
# Get all the flywire IDs (updated regularly based on position information) users have flagged +gs = googlesheets4::read_sheet(ss = options()$flywire_flagged_gsheet, sheet = "flywire") + +# Get IDs for User 'Tots' +gs.chosen = subset(gs, User == "Tots") + +# Load processed flywire neurons +fw = flywire_neurons() + +# Get those for Tots +fw.tots = fw[names(fw)%in%gs.chosen$flywire.id] + +# Examine their meta data +fw.tots[,]- + hemibrain and flywire meta data + +
- flywire
-
@@ -124,6 +127,9 @@
The best option is to use google filestream. By default, this is what
hemibrainrexpects. However, you need a Google Workspace account (formerly G-Suite), which is a paid-for service. Without this, the best option is to use rclone. You can also download to an external hard drive and use that.We have two Google team drives available for you to use, which contain similar data. One (
+"hemibrain") is for internal use by the Drosophila Connectomics Group. The other one ("hemibrainr") is shared with those who would like access. Contact us by email to request access.+ 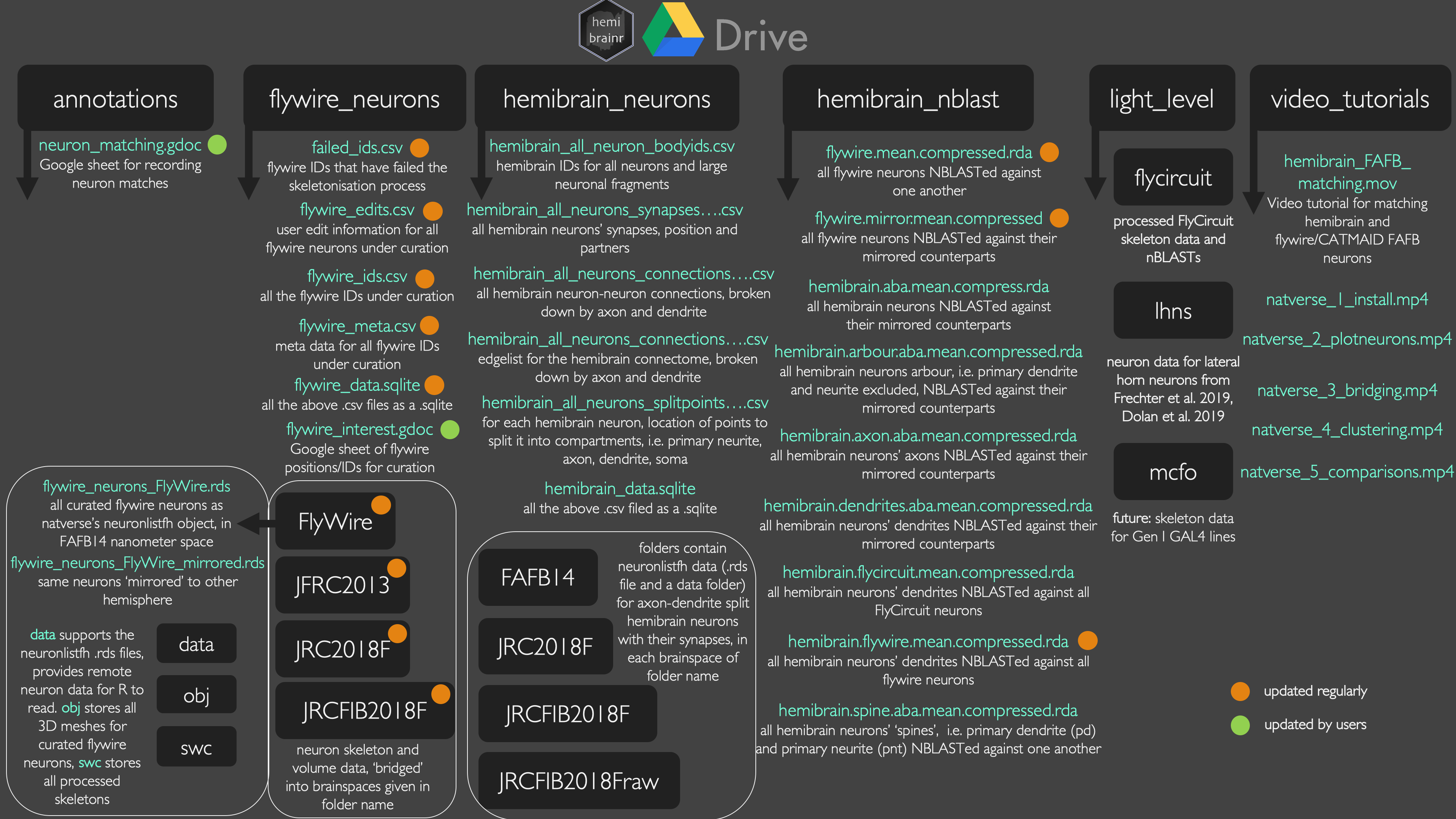 +@@ -194,7 +197,7 @@
+@@ -194,7 +197,7 @@Option 1: Google workspace, hemibrainr and R
@@ -162,7 +168,7 @@Set your drive location
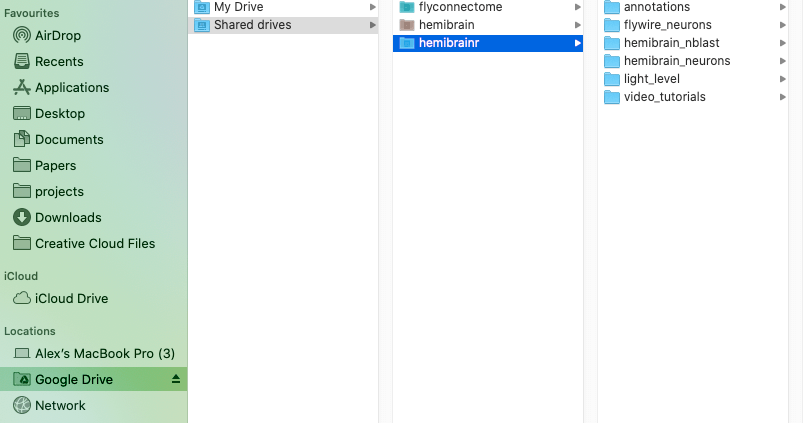
If you have mounted your Google drives with Google file stream, you should be able to see something like this:
- 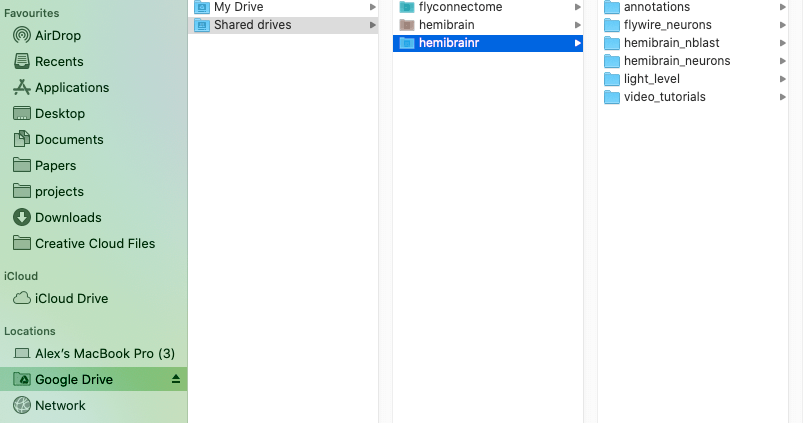 +
+
If you need to set
hemibrainrto look at a new drive, use:@@ -270,7 +276,7 @@
Once you have installed and configured rclone, you can use it to mount the google team drive. To do this, you create an empty directory wherever you want, named whatever you like, and then rclone ‘turns’ this directory into your mounted Google drive.
You can mount with rclone from your system’s command line:
+rclone mount hemibrainr: /path/to/local/mountTry it somewhere like
/Documentsto see how it works. Forhemibrainrthere is an R function that will do the mounting for you in your working directory. So within R:diff --git a/docs/articles/hemibrain_axons_dendrites.html b/docs/articles/hemibrain_axons_dendrites.html index a44a4b82..2e2d6d0c 100644 --- a/docs/articles/hemibrain_axons_dendrites.html +++ b/docs/articles/hemibrain_axons_dendrites.html @@ -55,6 +55,9 @@# mounts in working directory diff --git a/docs/articles/hemibrain_alpns_toons.html b/docs/articles/hemibrain_alpns_toons.html index ab067925..8b9a73c8 100644 --- a/docs/articles/hemibrain_alpns_toons.html +++ b/docs/articles/hemibrain_alpns_toons.html @@ -55,6 +55,9 @@- + hemibrain and flywire meta data + +
- flywire
-
@@ -161,12 +164,12 @@
Get neuron connectivity data
-Now we can get a precomputed edgelist for the brain from the Google drive, which breaks down connections by axon and dendrite. By default this is read from an SQLlite database on the Google drive. This means we can access the data, without loading a multi Gb .csv into memory:
+Now we can get a precomputed edgelist for the brain from the Google drive, which breaks down connections by axon and dendrite. By default this is read from an SQLite database on the Google drive. This means we can access the data, without loading a multi Gb .csv into memory:
-# Get the edgelist elist = hemibrain_elist() meta = rbind.fill(ton.info, pn.info)In our
+elist, count is the number of synaptic connections between two arbours. The arbour types are given byLabelandpartner.Label. The neuron identities are given aspre, the upstream (source) neuron andpost, the downstream (target) neuron. The value ofnormis generated by takingcountand dividing it by the total number of posynapses on the downstream target’s given arbour (note, not all connection in the neuron!). See your article on neuron splitting, for more information.In our
elist, count is the number of synaptic connections between two arbours. The arbour types are given byLabelandpartner.Label. The neuron identities are given aspre, the upstream (source) neuron andpost, the downstream (target) neuron. The value ofnormis generated by takingcountand dividing it by the total number of postsynapses on the downstream target’s given arbour (note, not all connection in the neuron!). See your article on neuron splitting, for more information.alpn.lhn.elist <- elist %>% filter(pre %in% local(pn.info$bodyid) @@ -199,7 +202,7 @@} conn.mat.scaled = t(apply(conn.mat,1,normalise))
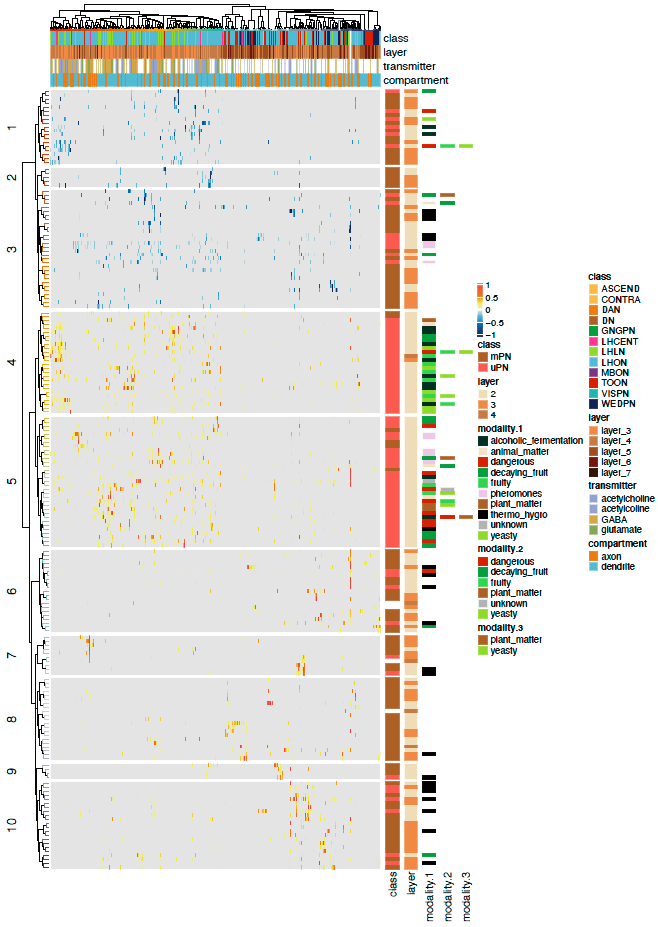
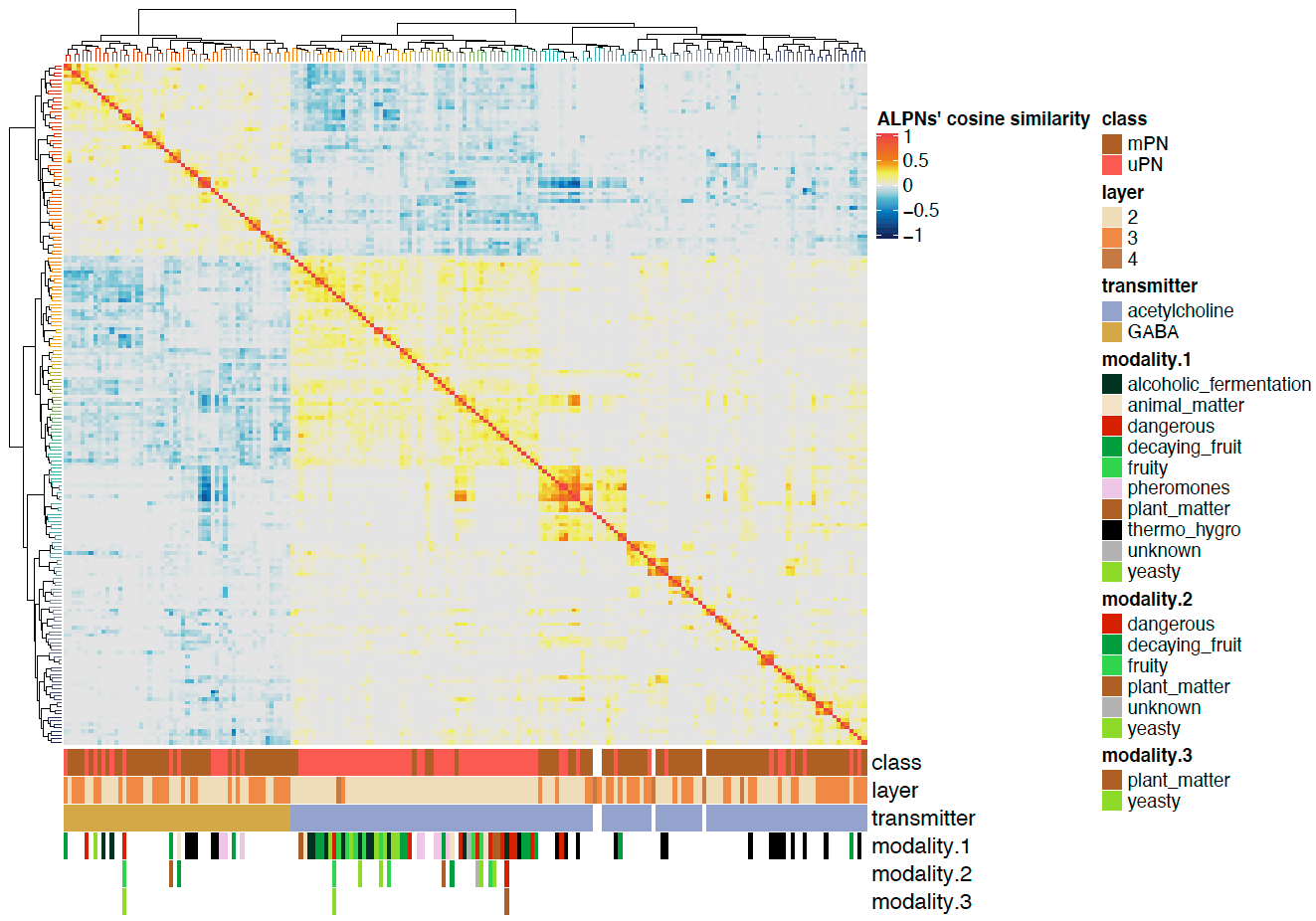
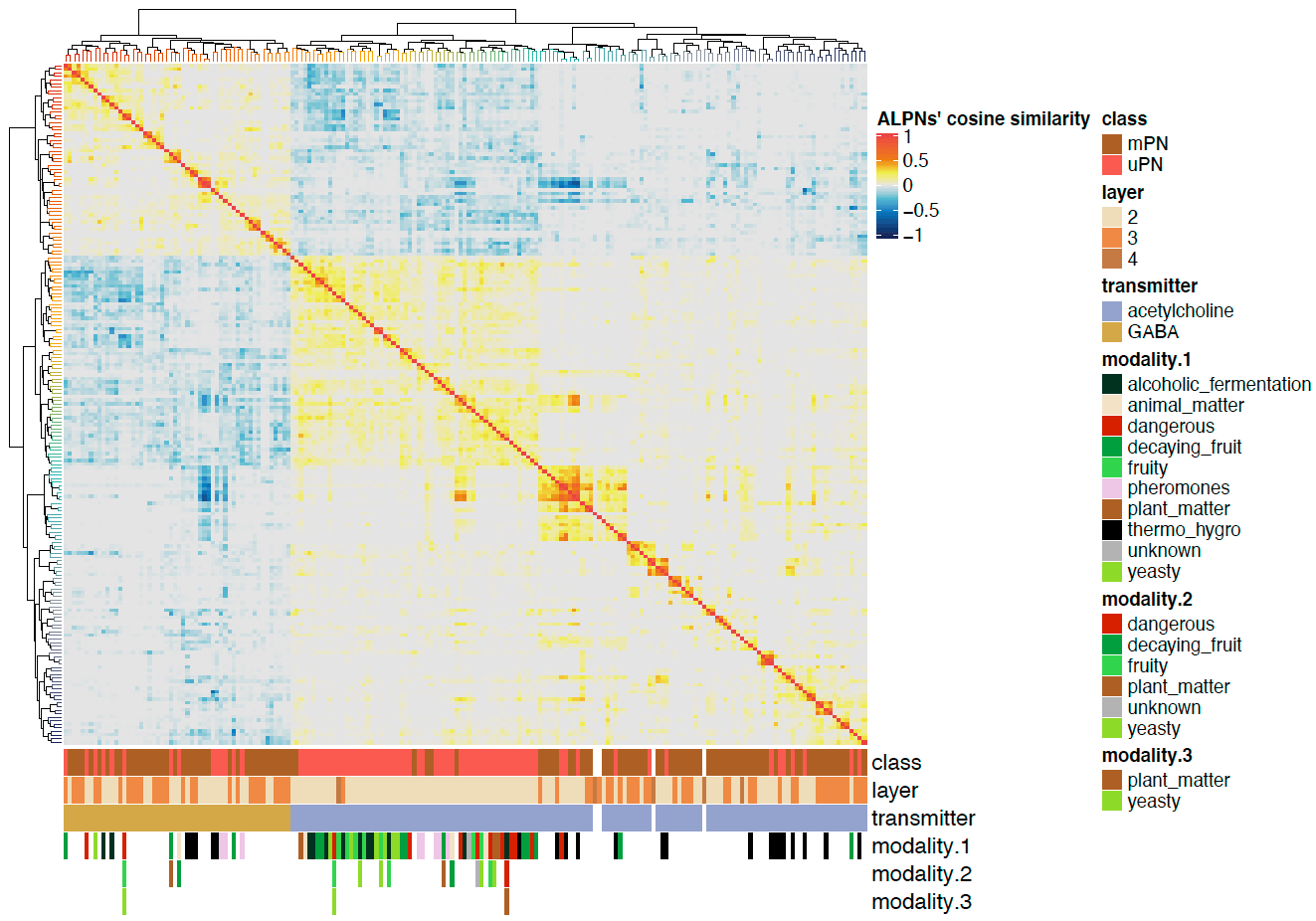
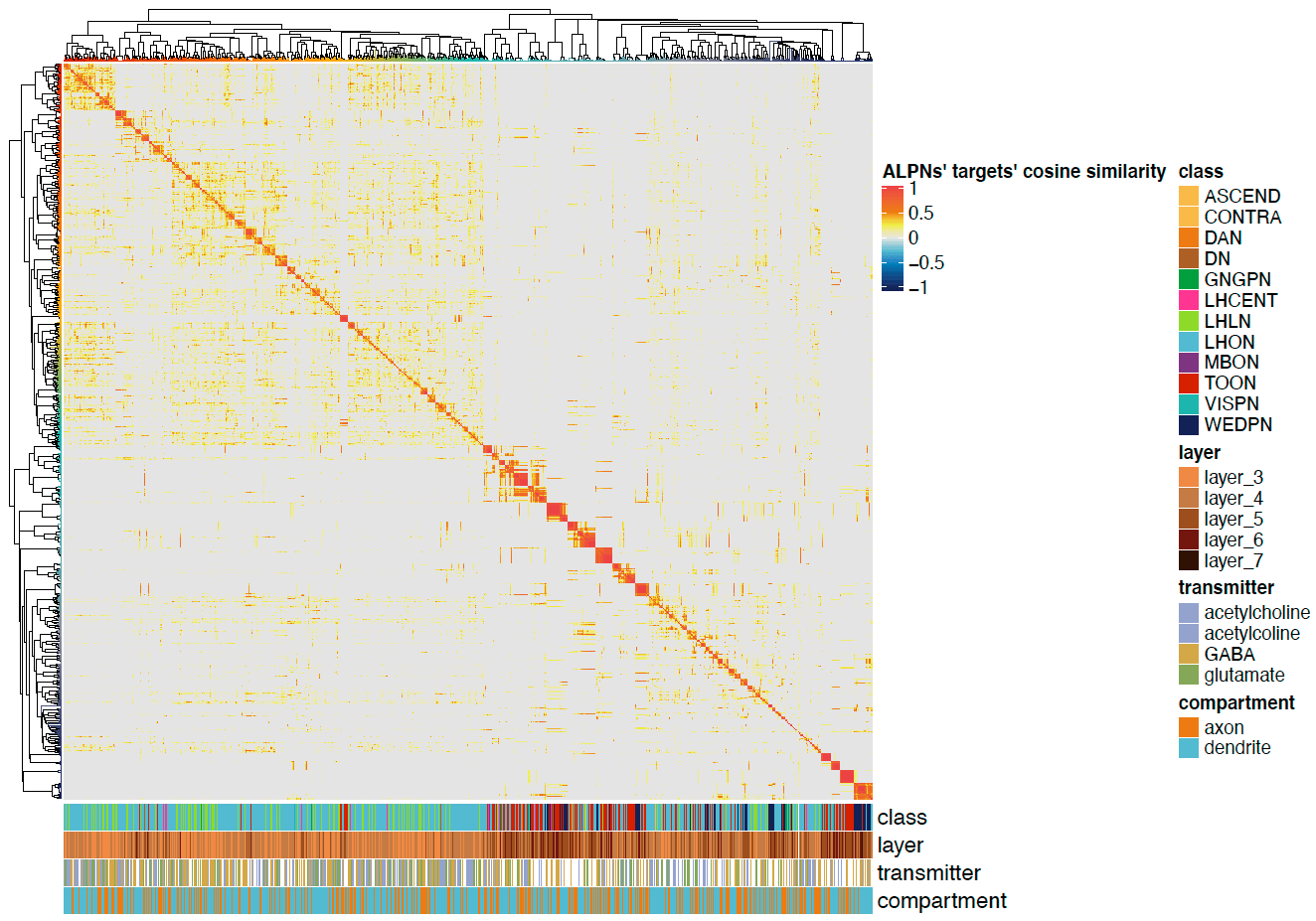
Now we can plot a heatmaps of the ALPN->TON weights, and correlation heatmaps.
-First let us get some clusterings for ALPNs and TONs based on ther cross-correlation.
+First let us get some clusterings for ALPNs and TONs based on their cross-correlation.
# TON correlation matrix cor.mat = lsa::cosine(conn.mat.scaled) @@ -340,7 +343,7 @@column_names_gp = gpar(fontsize = 12, fontfamily ="Helvetica")) dev.off()
- 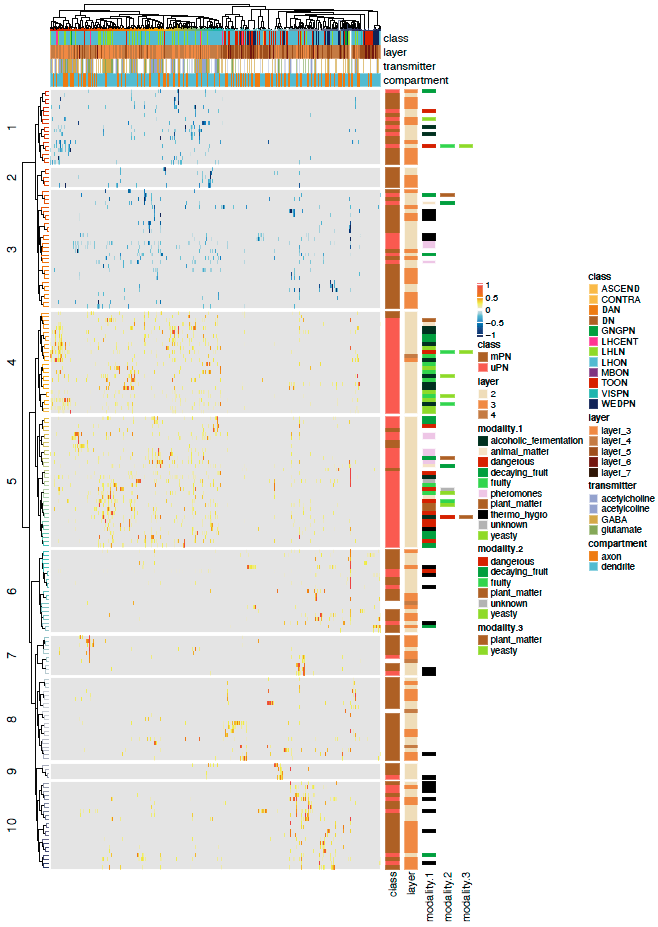 +
+
We can also plot the correlation heatmaps.
@@ -379,10 +382,10 @@
) dev.off()
-  +
+
-  +
+
- + hemibrain and flywire meta data + +
- flywire
-
@@ -121,12 +124,12 @@
Splitting a neuron
Neurons can be split into a principle input zone, a dendrite, and a principle output zone, an axon. Dendrites tend to be more spindly and axons bulkier, with boutons packed with presynapses, ready to release neurotransmitter.
- 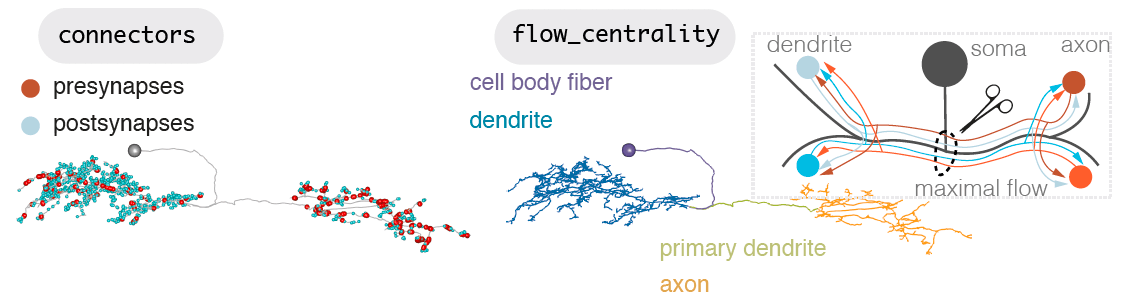 +
+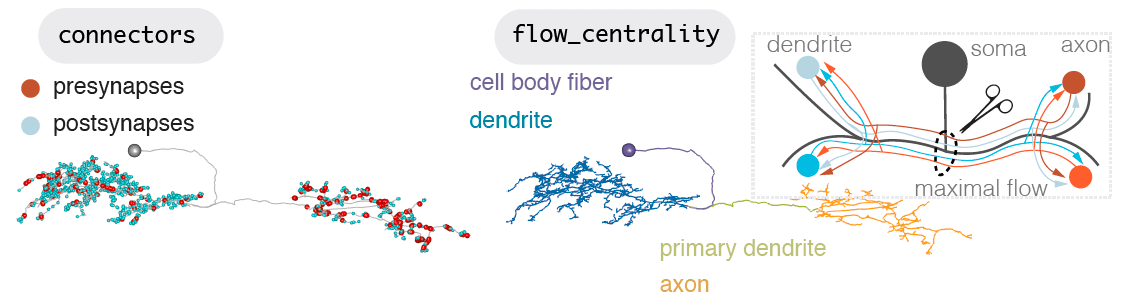
But axons and dendrites are not a totally clear division for most neurons. A neuron may have more than one of each. It may also be more ‘amacrine-like’ and lack this division, though this is less common. Dendrites can have outputs, and axons can have inputs. Axons may connect with axons, even dendrites may input and axon.
However, all fly neurons examined can be split into an axon-like and a dendrite-like compartment (Schneider-Mizell et al., 2016)). The density of outputs in the axon is higher than in the dendrite. The density of inputs to the dendrite, are higher than in the axon (Bates & Schlegel et al., 2020)). Interestingly, the ‘start’ of a dendrite is almost always closer to the neuron’s cell body than the start of its axon. See our recent data (Bates & Schlegel et al., 2020)) for three broad classes of olfactory neuron:
- 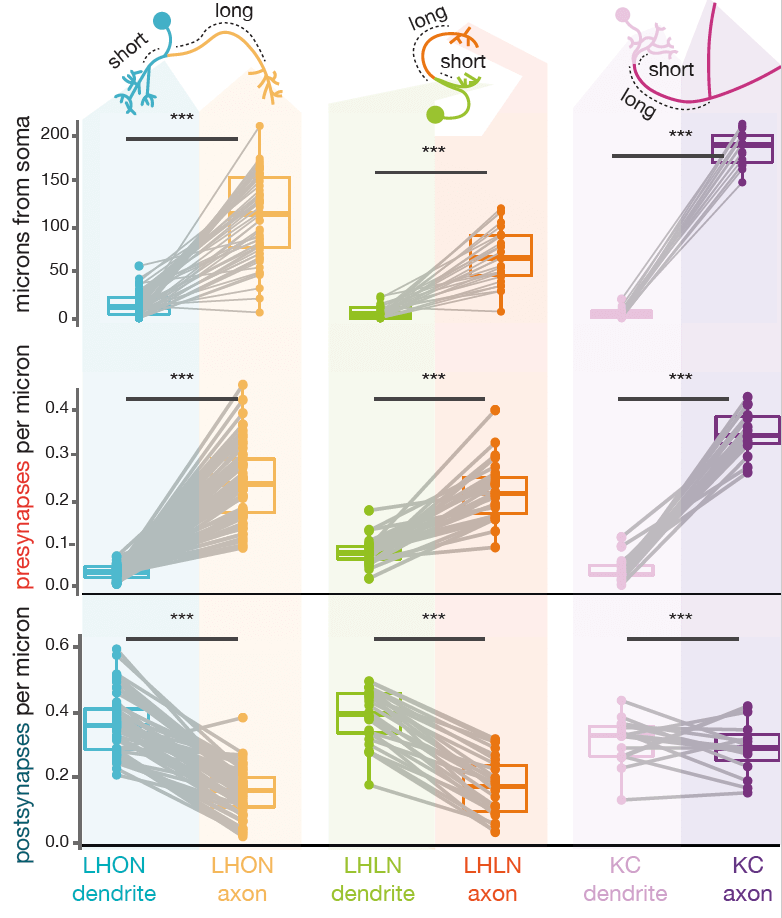 +
+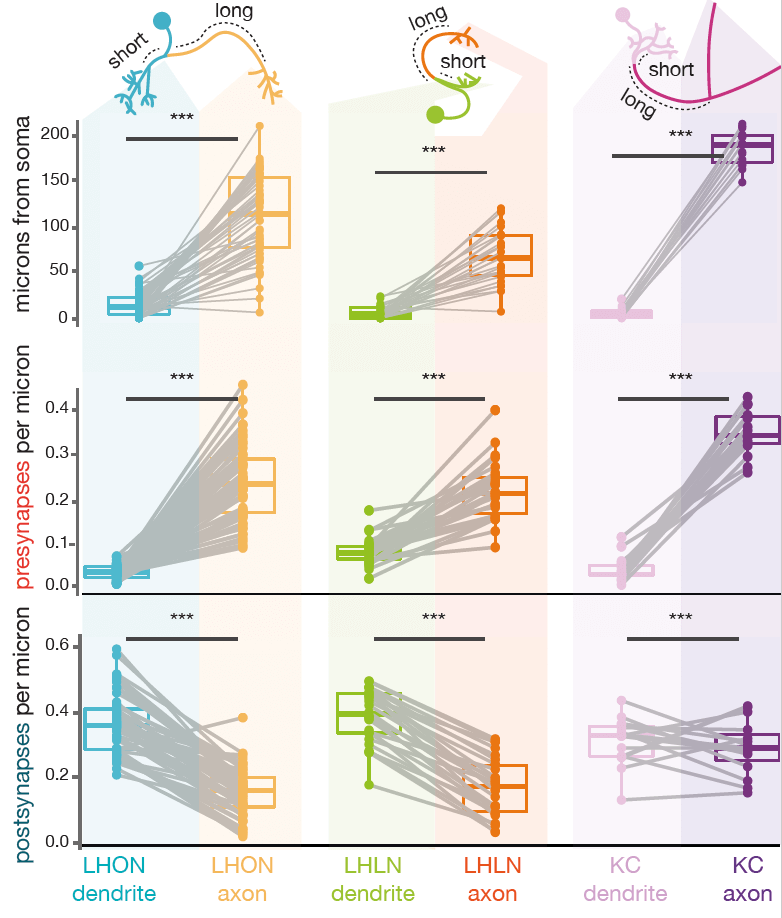
The biophysical impact of axo-axonic connections is unclear – they can have non-intuitive effects depending on the timing of action potentials in connected partners (Burrows and Laurent, 1993; Burrows and Matheson, 1994; Cuntz et al., 2007; Haag and Borst, 2004).
Get already split neurons
-Splitting neurons can take a little time. We can also load thousands of pre-split neurons from the
+hemibrainrGoogle team drive. In order to connect R to this Google drive, you have a few options. Please see this article. Once you have access and have mounted the drive, you should be able to do:Splitting neurons can take a little time. We can also load thousands of pre-split neurons from the
hemibrainrGoogle team drive. In order to connect R to this Google drive, you have a few options. Please see this article. Once you have access and have mounted the drive, you should be able to do:# All split hemibrain neurons db = hemibrain_neurons() @@ -234,7 +237,7 @@axon.synapses = hemibrain_extract_synapses(axons, prepost = "PRE") head(axon.synapses)
-  +
+ @@ -248,7 +251,7 @@
@@ -248,7 +251,7 @@plot3d_split(neurons.flow) plot3d(hemibrain.surf, col = "lightgrey", alpha = 0.1, add = TRUE)
-  +
+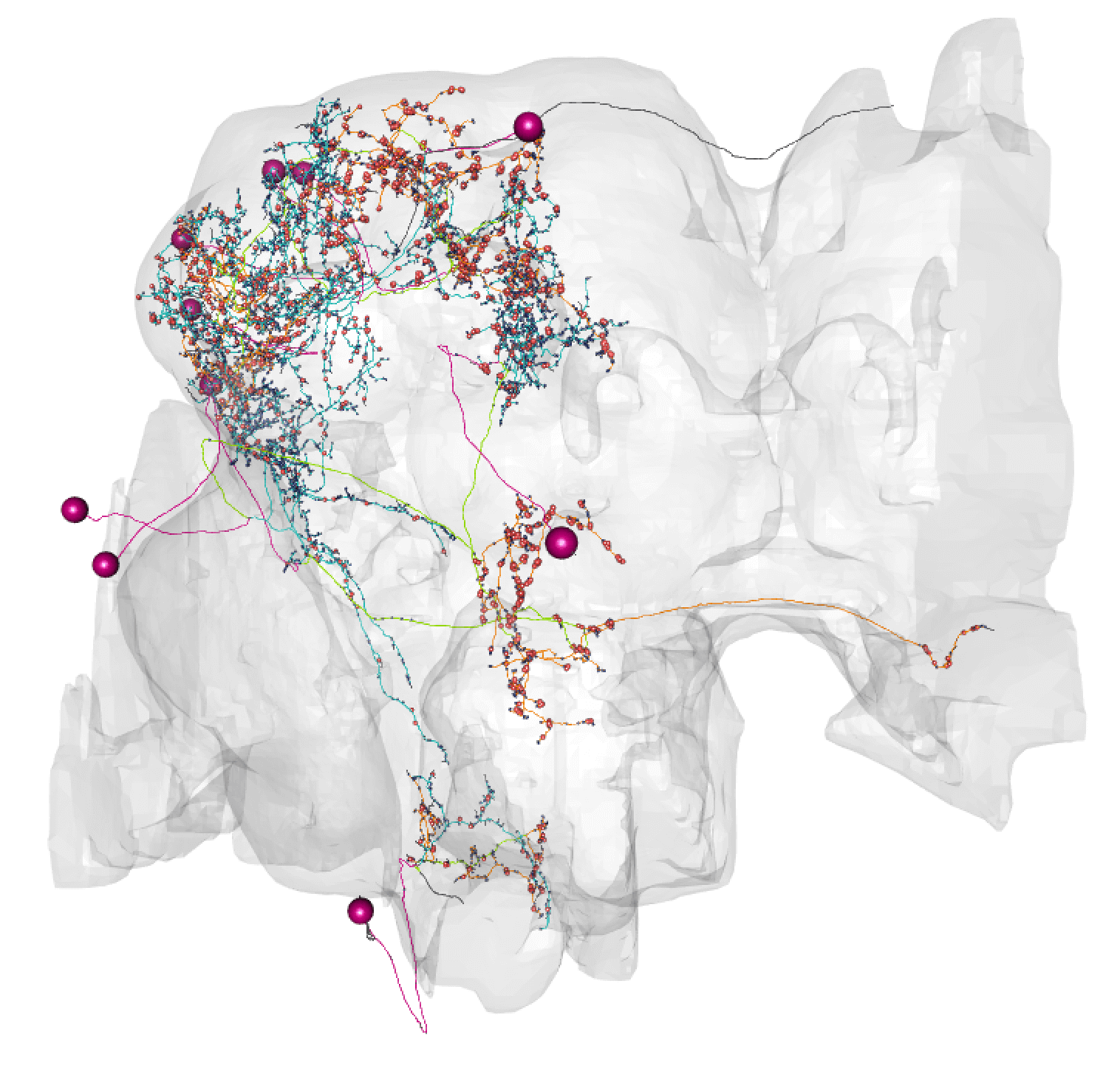
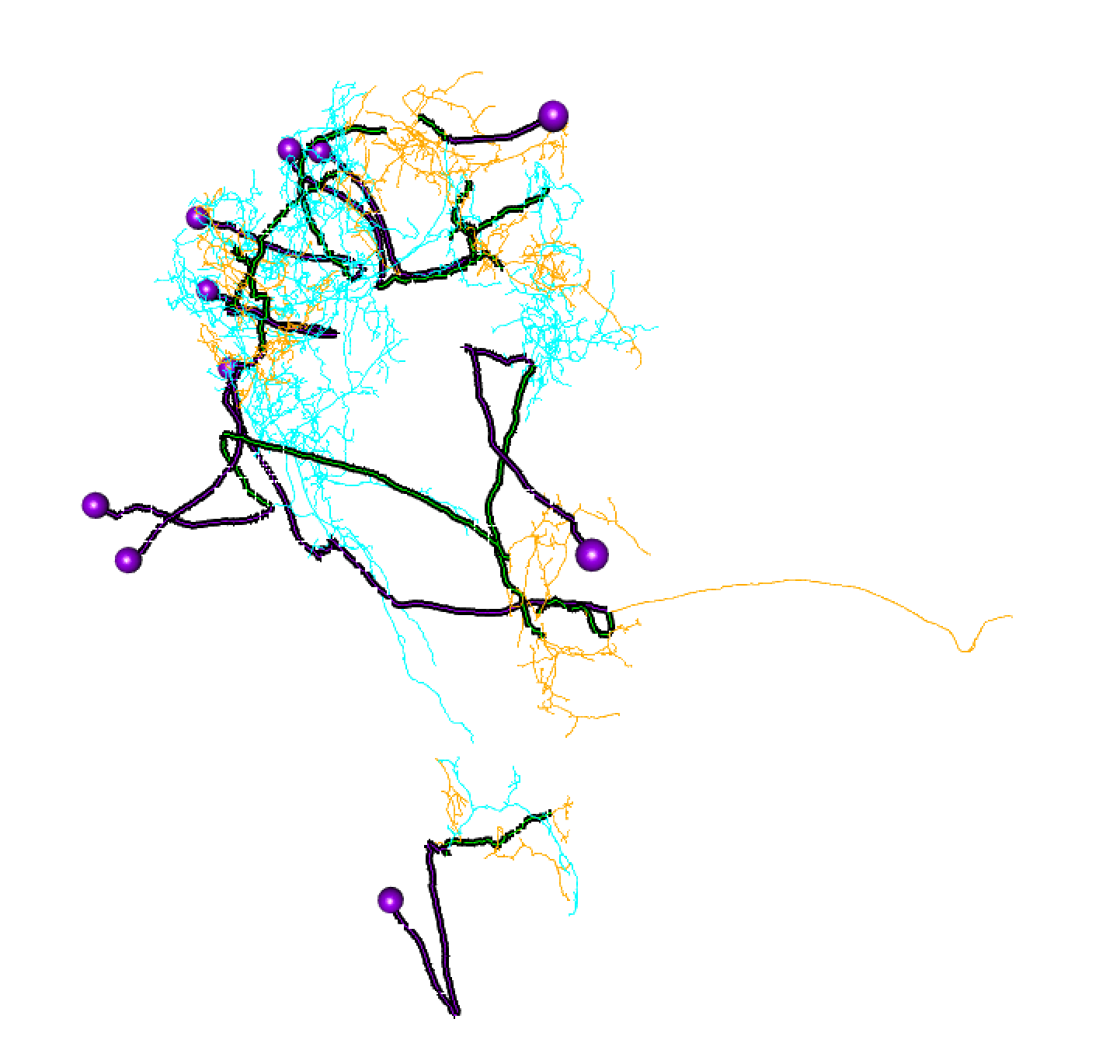
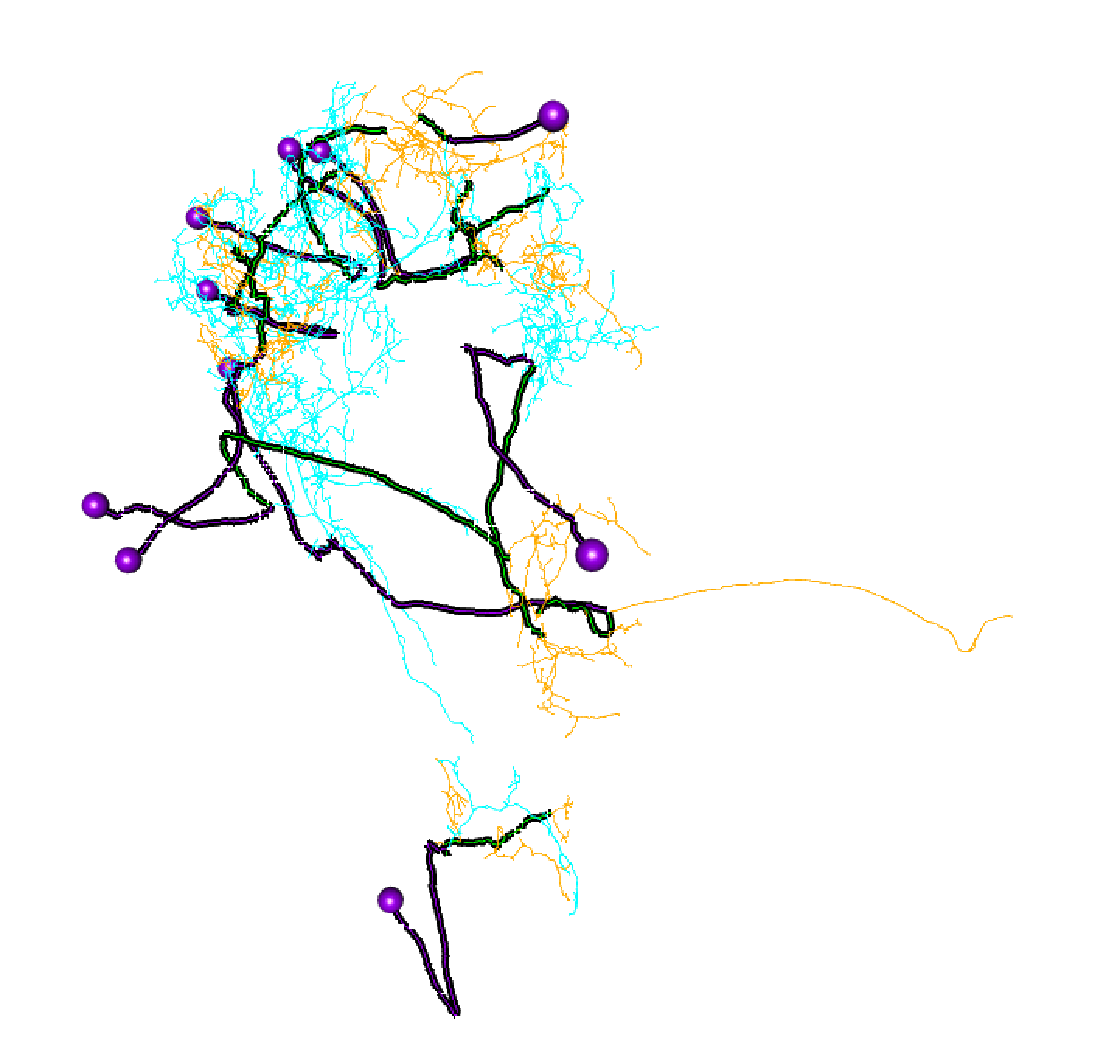
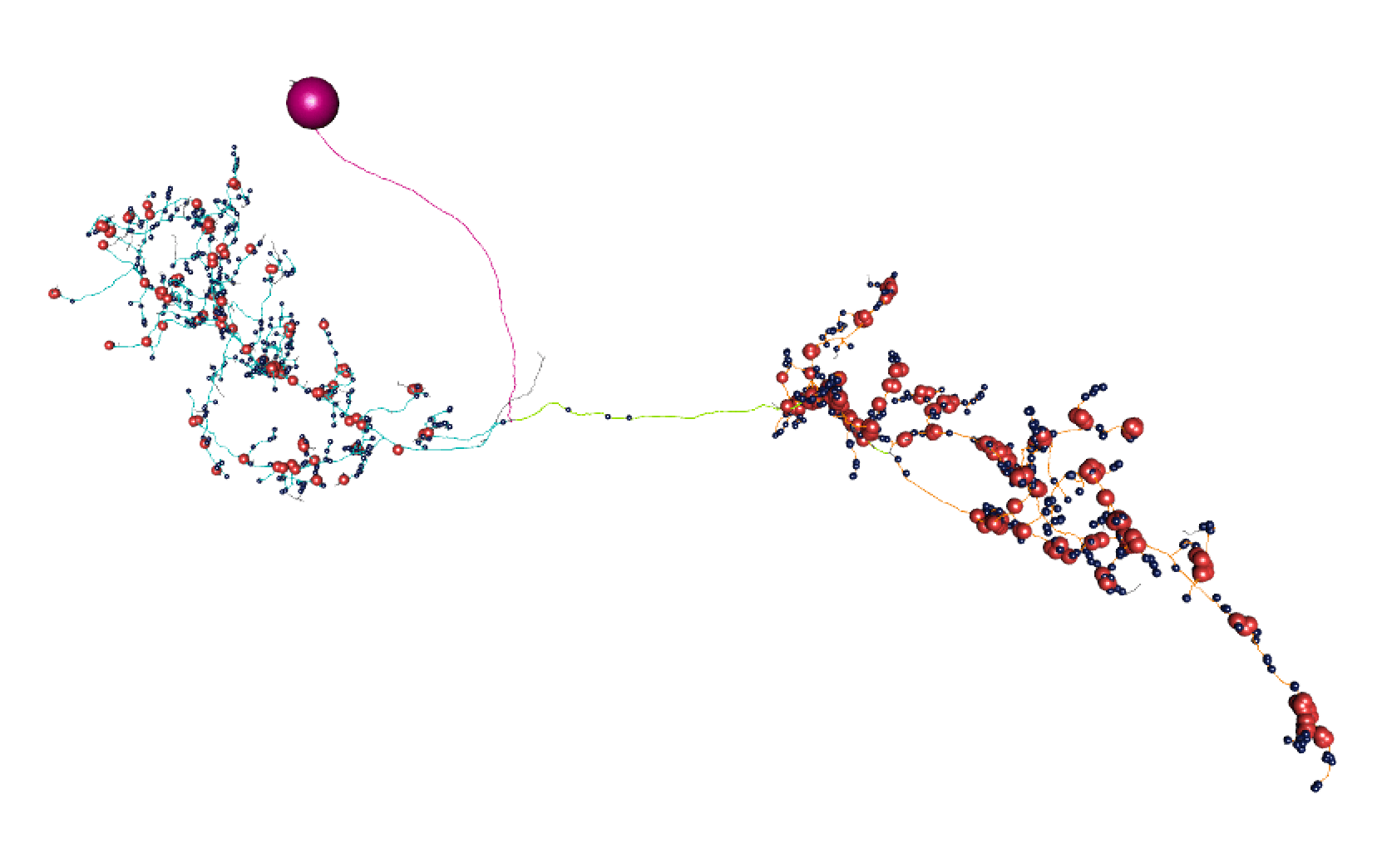
Here, lines: blue = dendrite, orange = axon, green = linker, purple = cell body fibre.
Balls: red = presynapse (output), navy blue = postsynapse (input), purple = soma.
@@ -258,7 +261,7 @@clear3d() nlscan_split(neurons.flow)
-  +
+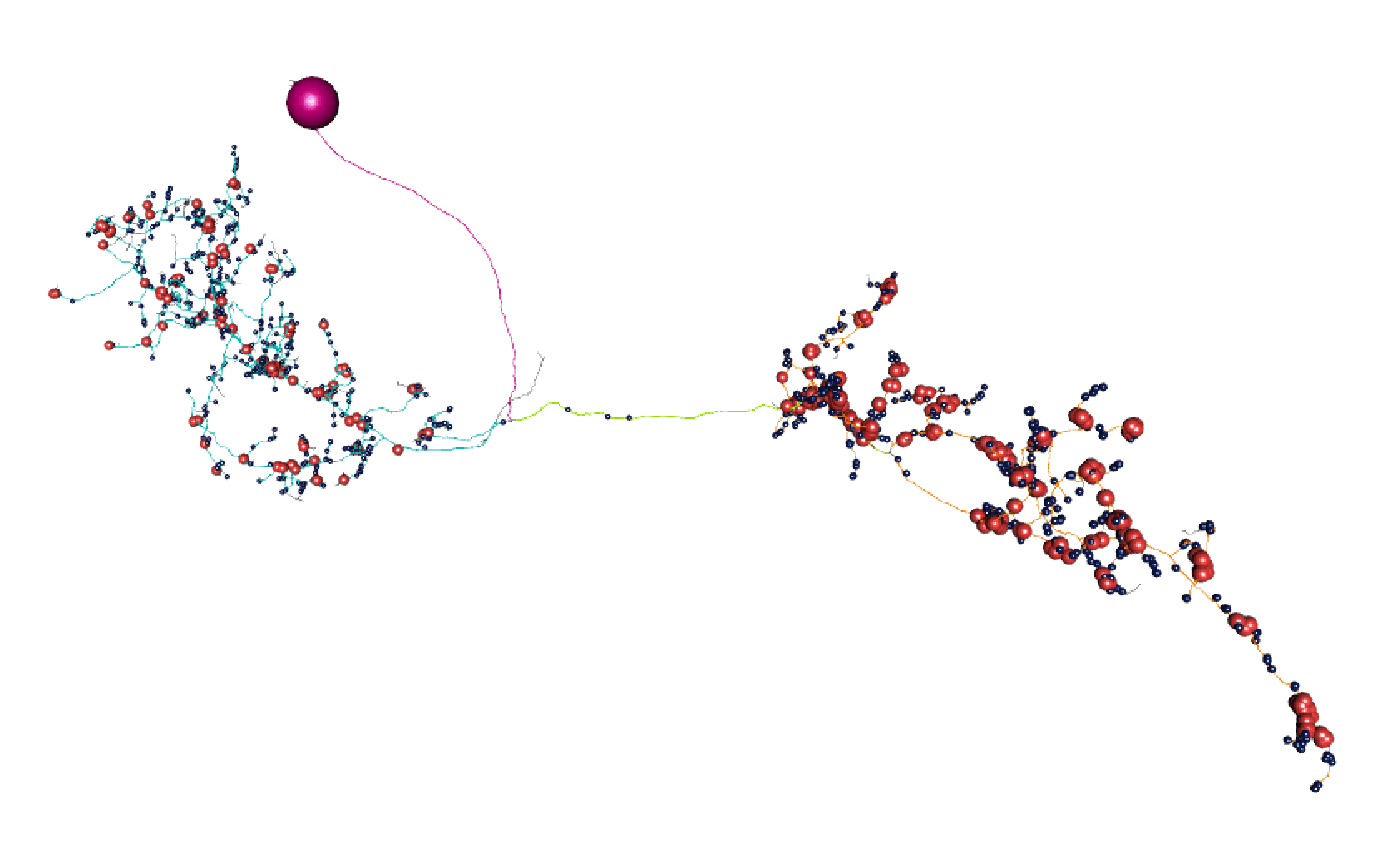
So I think most of those splits are pretty convincing.
@@ -310,7 +313,7 @@sort(table(pns.elist.strong$connection)) ## A lot of axo-axonic and dendro-dendritic action amongst the PNs
Here, pre is the
-bodyidof the presynaptic (upstream source) neuron and post is thebodyidof the postsynaptic (downstream target) neuron. Thenormcolumn gives a synaptic weight normalised by the total inputs of the downstream neuron (post), i.e. count/total postsynapses on ‘post’.Please see this article to see a more in-depth example of how to efficiently work neuron connectivity data from
+hemibrainr.Please see this article to see a more in-depth example of how to efficiently work neuron connectivity data from
diff --git a/docs/articles/hemibrain_connectivity.html b/docs/articles/hemibrain_connectivity.html index 279b40d9..8912b974 100644 --- a/docs/articles/hemibrain_connectivity.html +++ b/docs/articles/hemibrain_connectivity.html @@ -55,6 +55,9 @@hemibrainr.- + hemibrain and flywire meta data + +
- flywire
-
@@ -114,8 +117,8 @@
hemibrain connectivity
Connection data
-In this guide, we will use
-hemibrainrlook at connectivity data for a starting neuron of interest. They neuron will be LHMB1 (Bates & Schlegel et al., 2020)), the only neuron from the ‘innate olfactory’ brain center of the fly (lateral horn), to innervate the ‘associative memroy centre’ directly (mushroom body lobes). You can try re-running this analysis for any neuron of your interest.We will use precomputed data from the
+hemibrainrGoogle team drive. In order to connect R to this Google drive, you have a few options. Please see this article. To see how to split neurons into axon and dendrite, without acces to this drive, please see this article.In this guide, we will use
+hemibrainrlook at connectivity data for a starting neuron of interest. They neuron will be LHMB1 (Bates & Schlegel et al., 2020)), the only neuron from the ‘innate olfactory’ brain center of the fly (lateral horn), to innervate the ‘associative memory centre’ directly (mushroom body lobes). You can try re-running this analysis for any neuron of your interest.We will use precomputed data from the
hemibrainrGoogle team drive. In order to connect R to this Google drive, you have a few options. Please see this article. To see how to split neurons into axon and dendrite, without access to this drive, please see this article.Load google drive
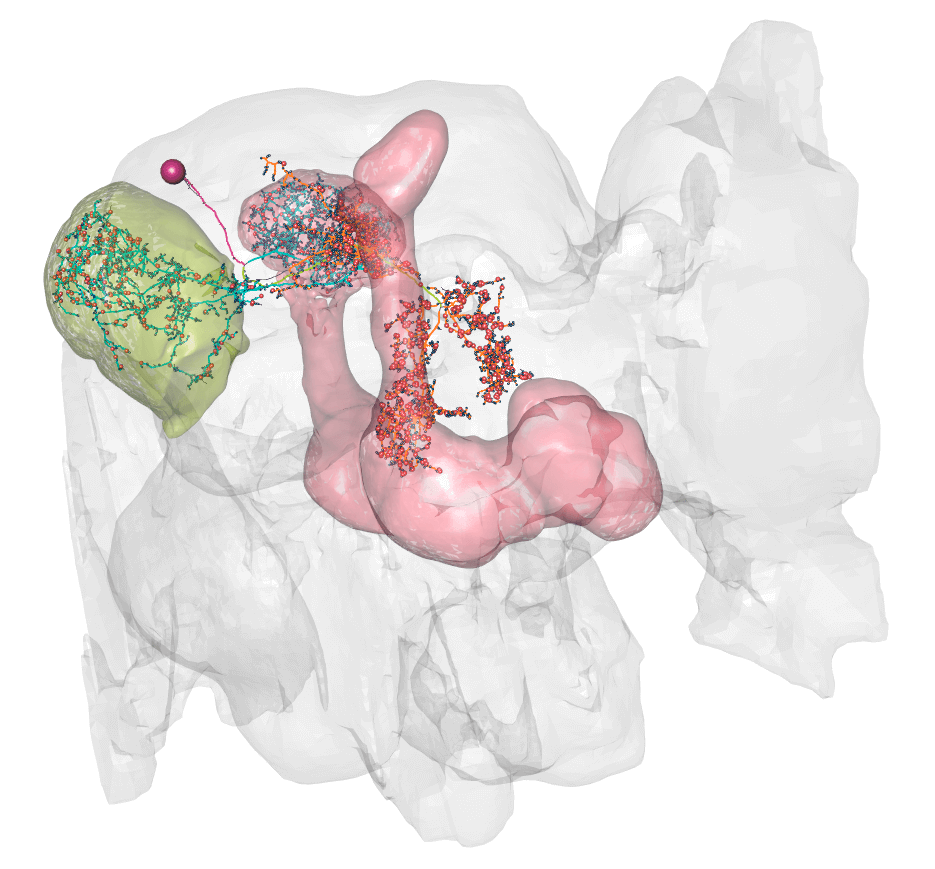
@@ -161,14 +164,14 @@plot3d(hemibrain.surf, alpha = 0.1, col = "lightgrey", add = TRUE) plot3d_split(lhmb1.neuron, lwd =2, soma = 600)
- - +
+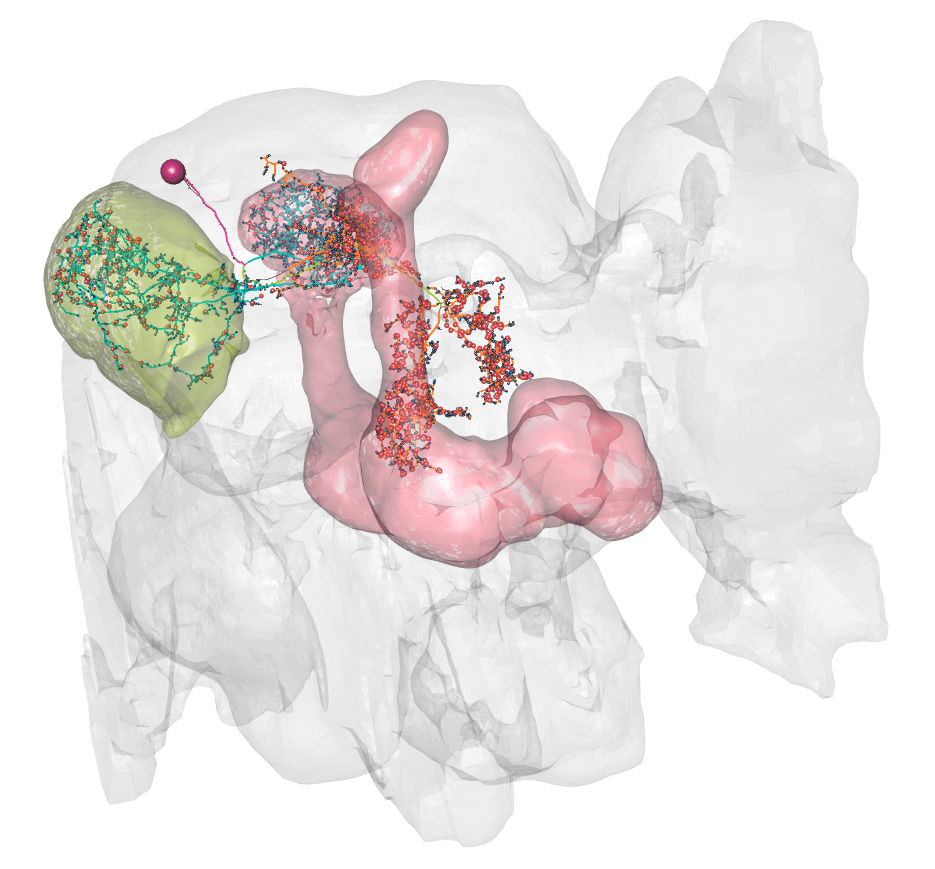
So here you can see dendrite (blue) in the lateral horn (green volume) and calyx, and axon (orange) in the lobes of the msuhroom body (pink volume).
+So here you can see dendrite (blue) in the lateral horn (green volume) and calyx, and axon (orange) in the lobes of the mushroom body (pink volume).
Get neuron connectivity data
-Now we can get a precomputed edgelist for the brain from the Google drive, which breaks down connections by axon and dendrite. By default this is read from an SQLlite database on the Google drive. This means we can access the data, without loading a multi Gb .csv into memory:
+Now we can get a precomputed edgelist for the brain from the Google drive, which breaks down connections by axon and dendrite. By default this is read from an SQLite database on the Google drive. This means we can access the data, without loading a multi Gb .csv into memory:
-# Get the edgelist elist = hemibrain_elist() @@ -178,8 +181,8 @@# Get a lot of meyta data for each hemibrain neuron hemi.meta = hemibrain_meta()
In our
-elist, count is the number of synaptic connections between two arbours. The arbour types are given byLabelandpartner.Label. The neuron identities are given aspre, the upstream (source) neuron andpost, the downstream (target) neuron. The value ofnormis generated by takingcountand dividing it by the total number of posynapses on the downstream target’s given arbour (note, not all connection in the neuron!). See your article on neuron splitting, for more information.We have the connectivity of whe whole dataset at our fingertips. Or at least, all the large fragments that could be considered neurons (see
+?hemibrain_bodyids). Let us have a quick look at what rthe distribution of different connection types is:In our
+elist, count is the number of synaptic connections between two arbours. The arbour types are given byLabelandpartner.Label. The neuron identities are given aspre, the upstream (source) neuron andpost, the downstream (target) neuron. The value ofnormis generated by takingcountand dividing it by the total number of postsynapses on the downstream target’s given arbour (note, not all connection in the neuron!). See your article on neuron splitting, for more information.We have the connectivity of the whole dataset at our fingertips. Or at least, all the large fragments that could be considered neurons (see
?hemibrain_bodyids). Let us have a quick look at what the distribution of different connection types is:
@@ -204,12 +205,11 @@lhmb1.elist <- elist %>% dplyr::filter(pre == lhmb1.bodyid | post == lhmb1.bodyid, count > 10 | norm > 0.01) %>% diff --git a/docs/articles/index.html b/docs/articles/index.html index b99a6189..6af7131e 100644 --- a/docs/articles/index.html +++ b/docs/articles/index.html @@ -95,6 +95,9 @@-
+
- + hemibrain and flywire meta data +
- flywire @@ -147,6 +150,8 @@
- hemibrain and flywire meta data +
- flywire
- read data from Google drive diff --git a/docs/articles/match_making.html b/docs/articles/match_making.html index cf2ec7d0..aab3733d 100644 --- a/docs/articles/match_making.html +++ b/docs/articles/match_making.html @@ -55,6 +55,9 @@
- + hemibrain and flywire meta data + +
- flywire
-
@@ -116,42 +119,42 @@
Neuron Matching Making
Insect brains seem pretty stereotyped. But just how stereotyped are they? It comes as a surprise to many neuroscientists who work only on vertebrates, to learn that in insects, individual neurons can readily and reliably be re-found and identified across different members of the species. Perhaps even across species.
-  +
+
As of 2020, two large data sets for the vinegar fly, D. melanogaster, are available making it possible to look at the full morphology of ~25,000 neurons in two data sets. These data sets are the hemibrain and FAFB. However, neurons in FAFB have been semi-manually or manually reconstructed, making the automatic assignment of FAFB-hemibrain neuron matches non-trivial. In this R package we have built tools to enable users to record and deploy inter-dataset matches.
What use is this information? Matches could be used to look at morphological stereotypy, help find genetic lines that label neurons, help transfer information associated with on reconstructed to the same cell in a different brain, compare neuron connectivity between two brains, etc.
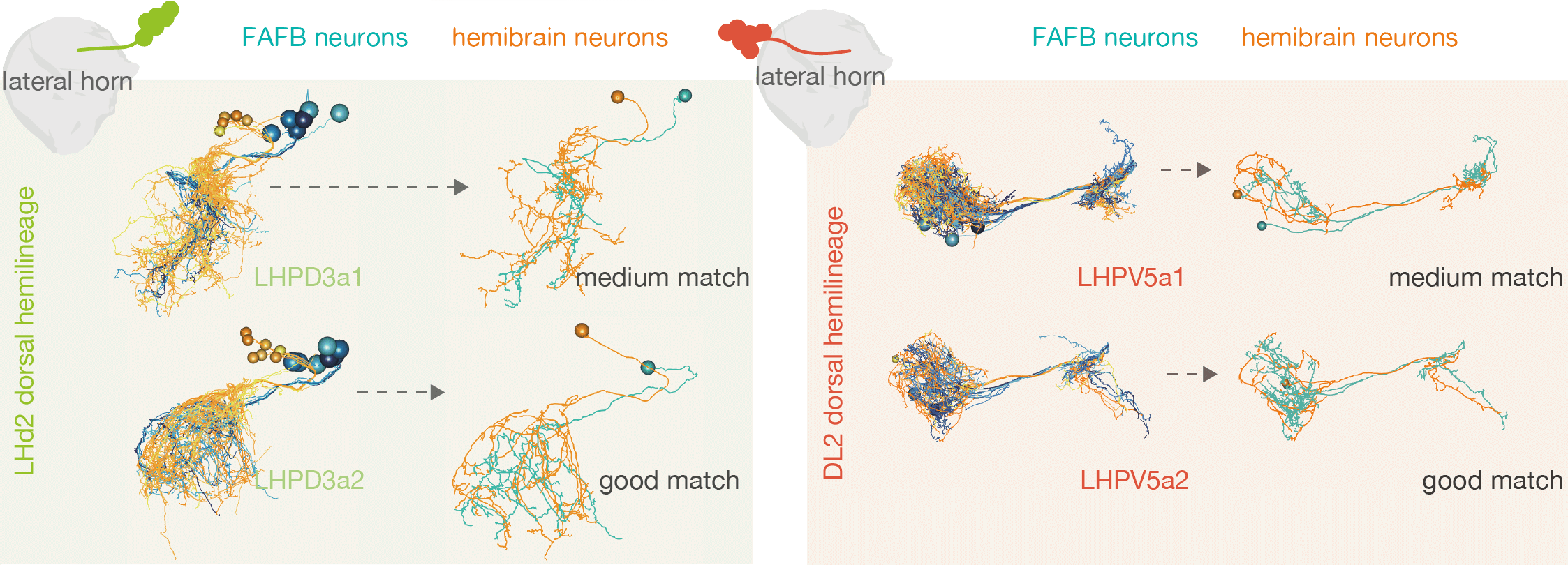
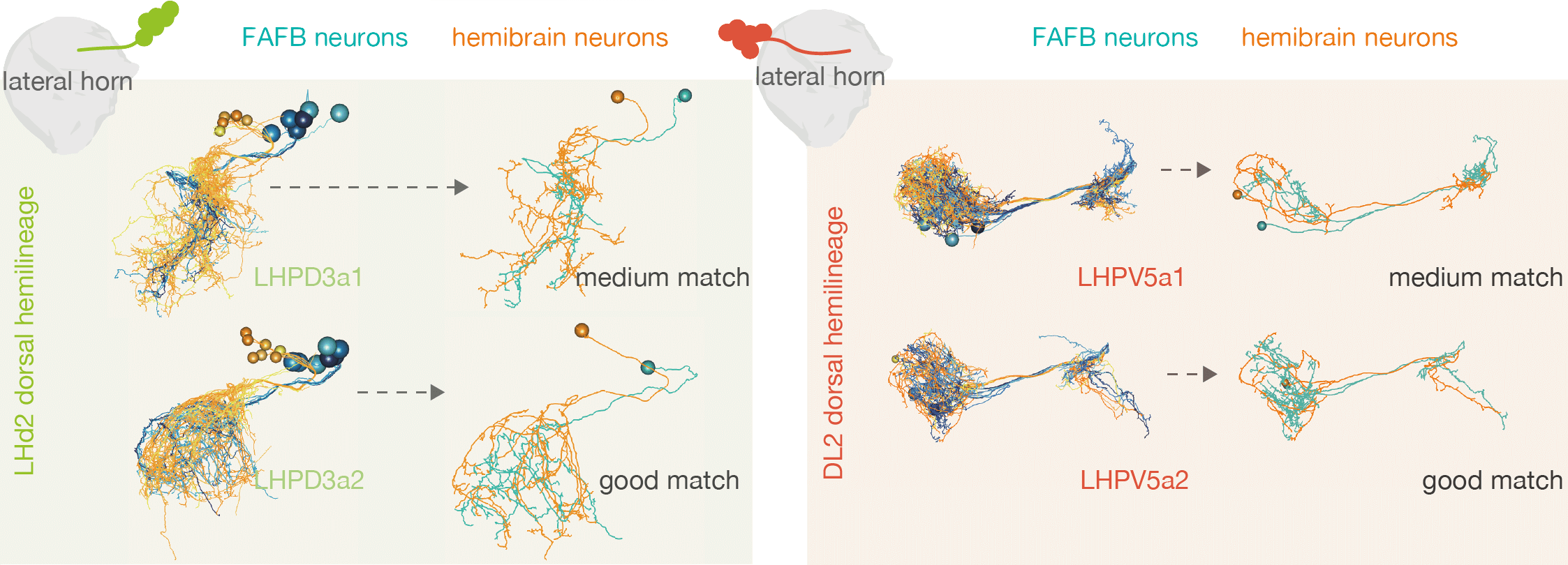
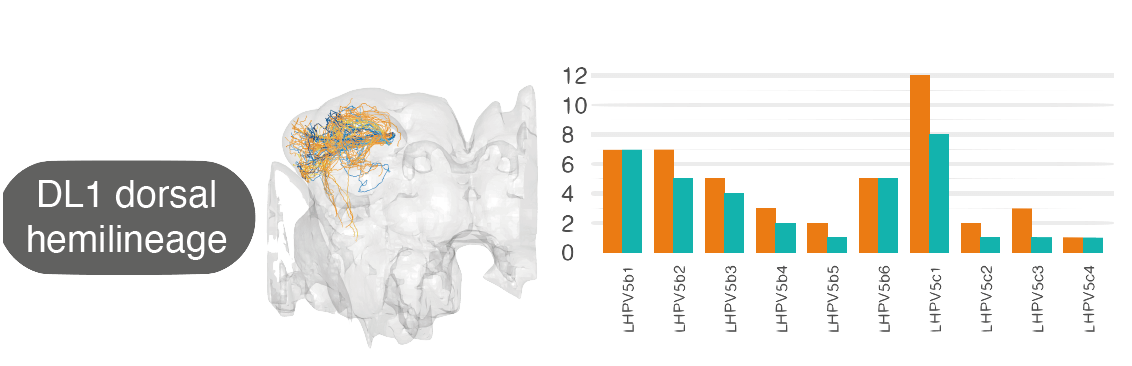
For example, by matching neurons up between the hemibrain and FAFB, we see that the numbers of cell types within one ‘hemilineage’ (a set of neurons that are born and develop together) are comparable between these two different flies:
- 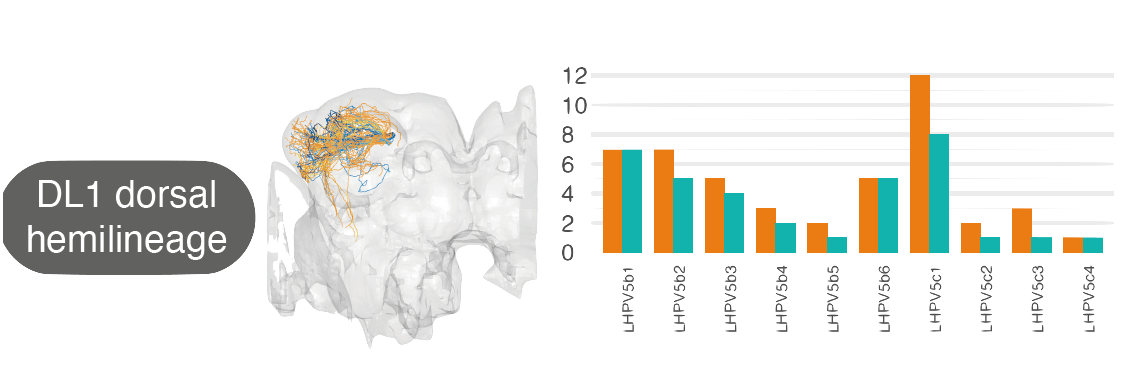 +
+
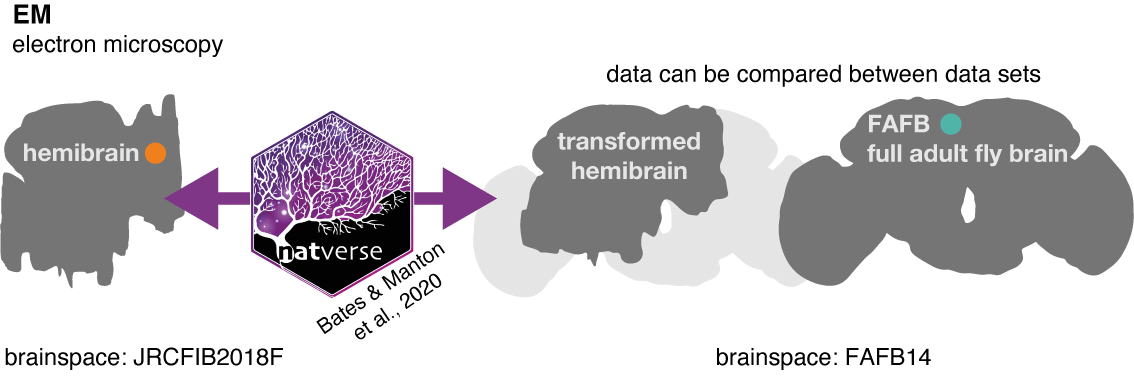
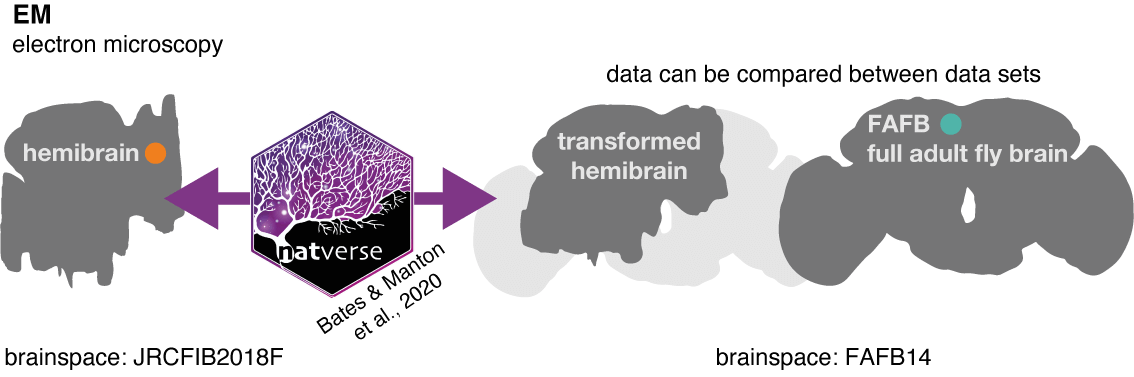
In order to match neurons, we make use of other natverse tools to ‘bridge’ data between two different brainspace, so they can be co-visualised (enabled by template brain and bridging registrations by Bogovic et al. 2019):
-  +
+
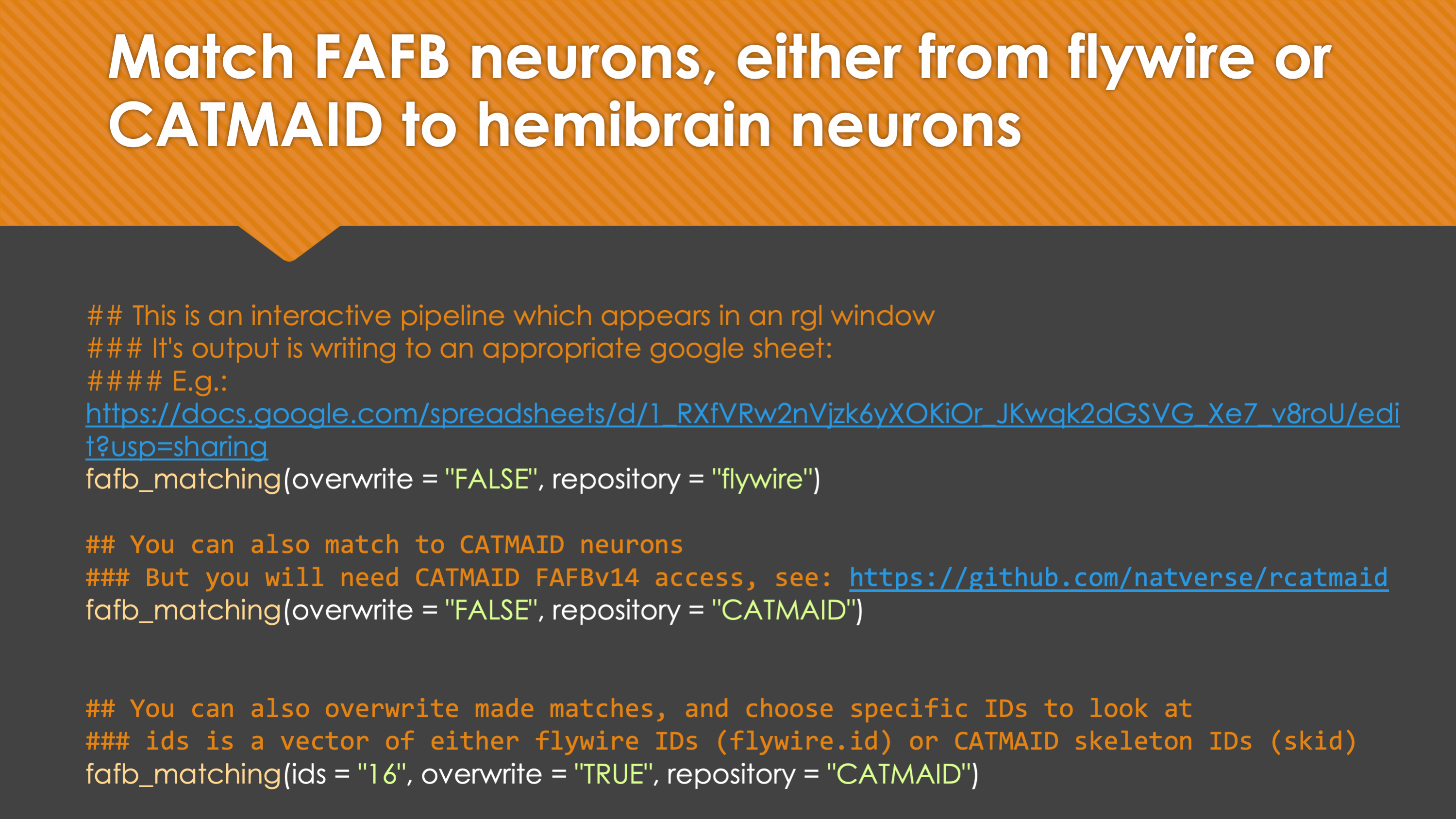
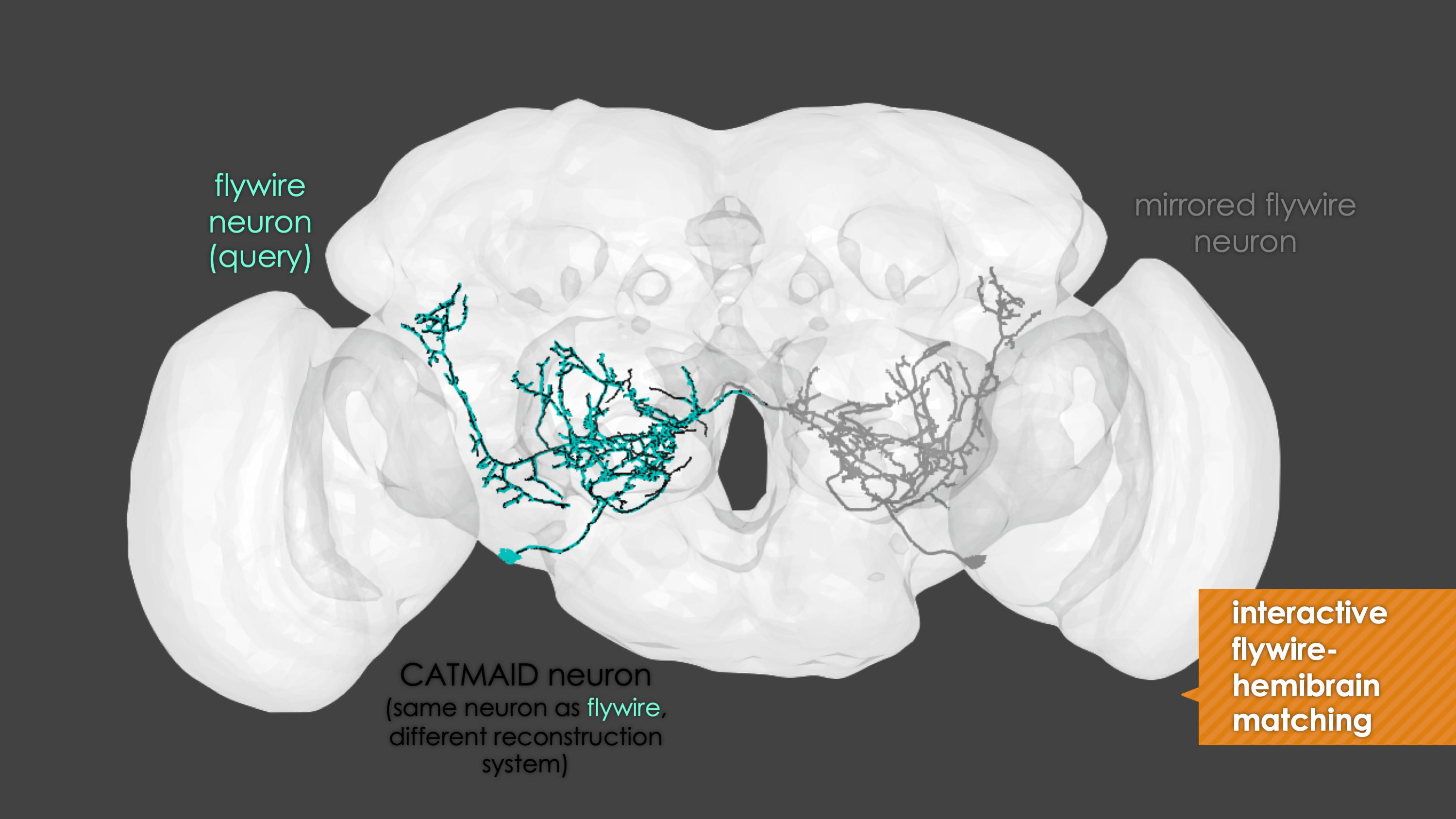
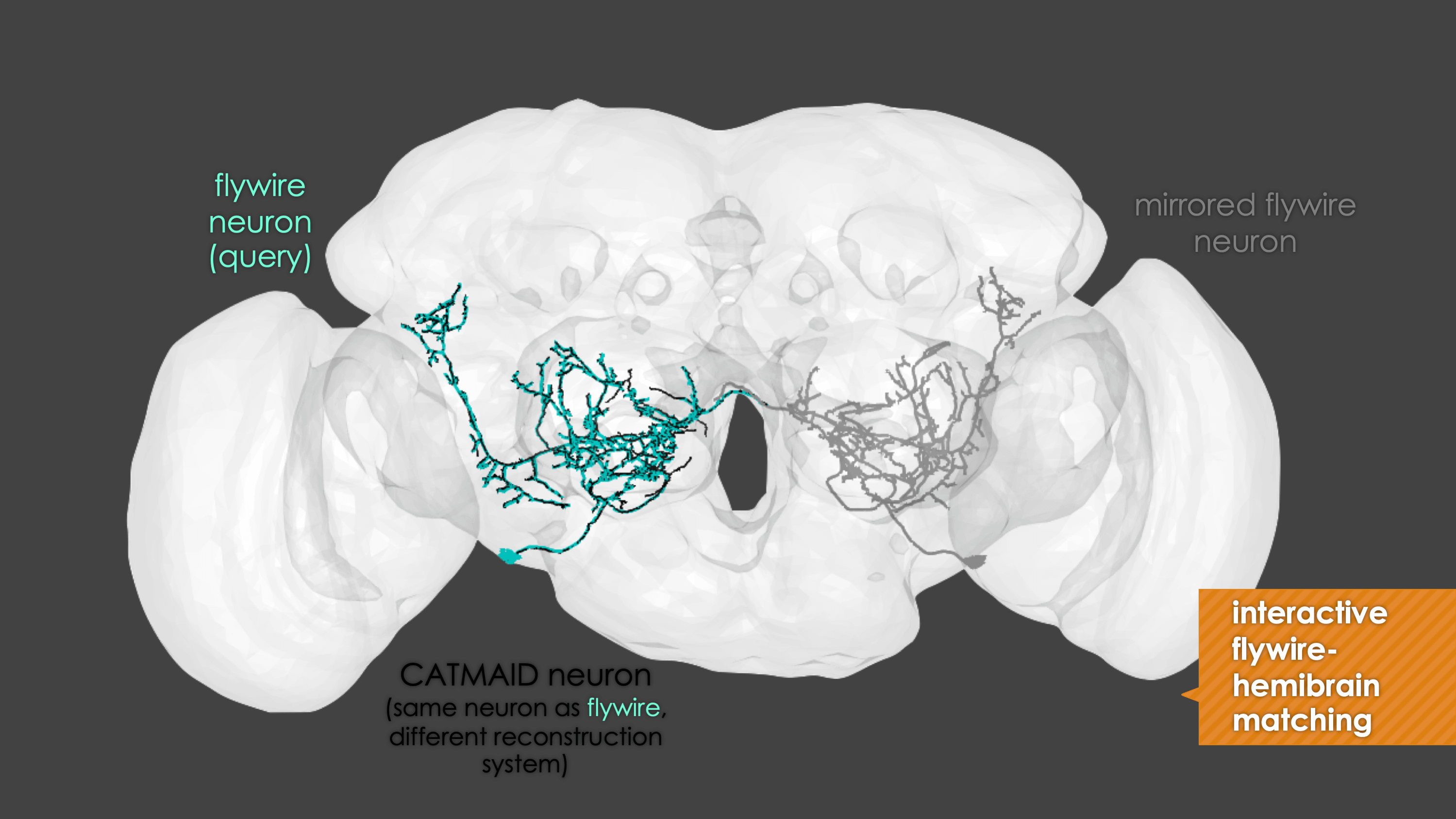
Overview
-In general, our interactive matching pipelines follow this workslow (this exmple is for
+fafb_matching):In general, our interactive matching pipelines follow this workflow (this example is for
fafb_matching):- 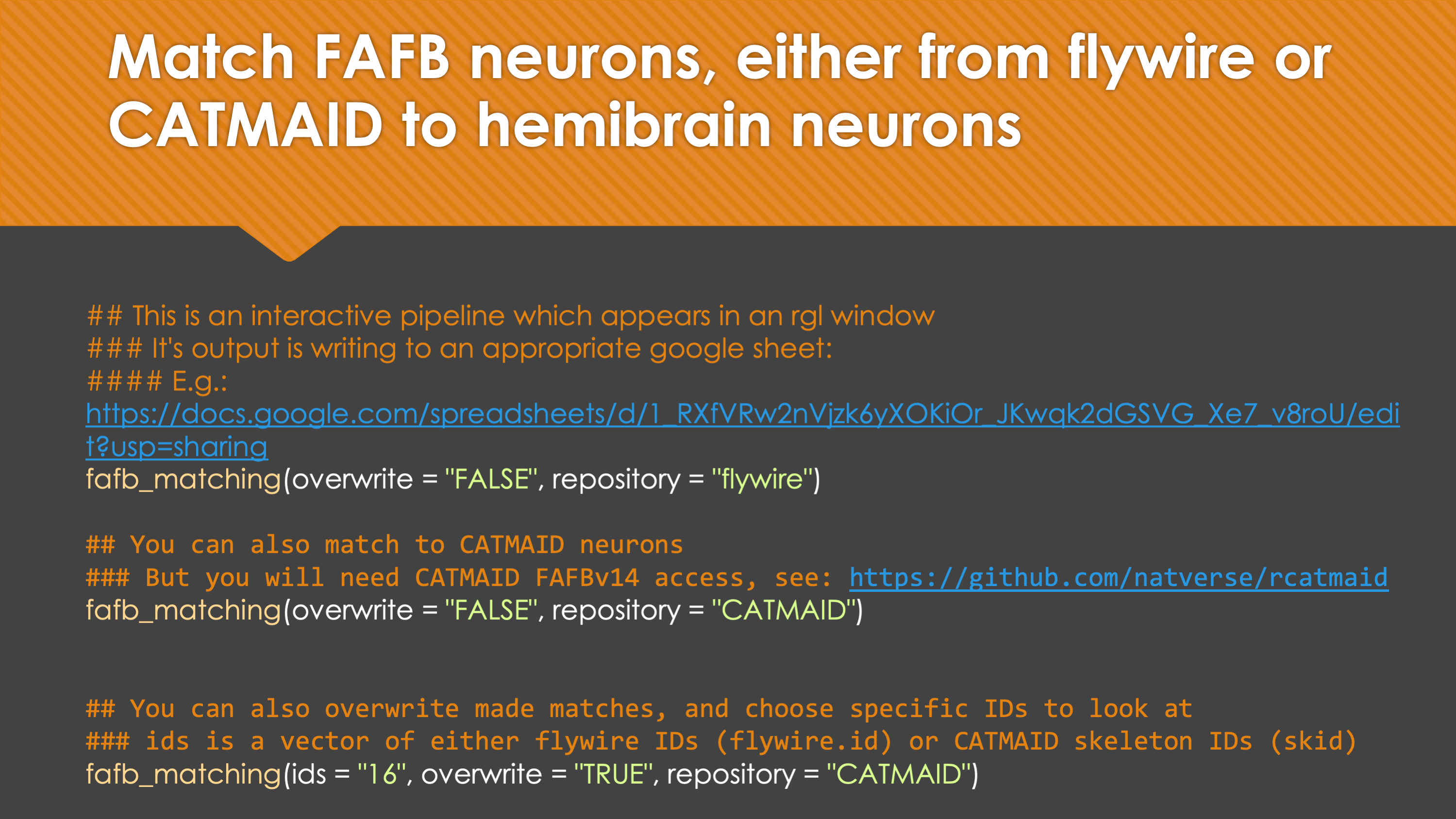 +
+
-  +
+
-  +
+
-  +
+
-  +
+
-  +
+
-  +
+ @@ -174,7 +177,7 @@diff --git a/docs/articles/packages_guide.html b/docs/articles/packages_guide.html index 23e9905c..10a81533 100644 --- a/docs/articles/packages_guide.html +++ b/docs/articles/packages_guide.html @@ -55,6 +55,9 @@
@@ -174,7 +177,7 @@diff --git a/docs/articles/packages_guide.html b/docs/articles/packages_guide.html index 23e9905c..10a81533 100644 --- a/docs/articles/packages_guide.html +++ b/docs/articles/packages_guide.html @@ -55,6 +55,9 @@The Google Sheet
We in the Drosophila Connectomics Group have been recording our match making in a Google sheet named em_matching. This sheet has two tabs of concern here,
hemibrainfor hemibrain neuron -> FAFB neuron matches andfafbfor FAFB neuron to hemibrain neuron matches.- 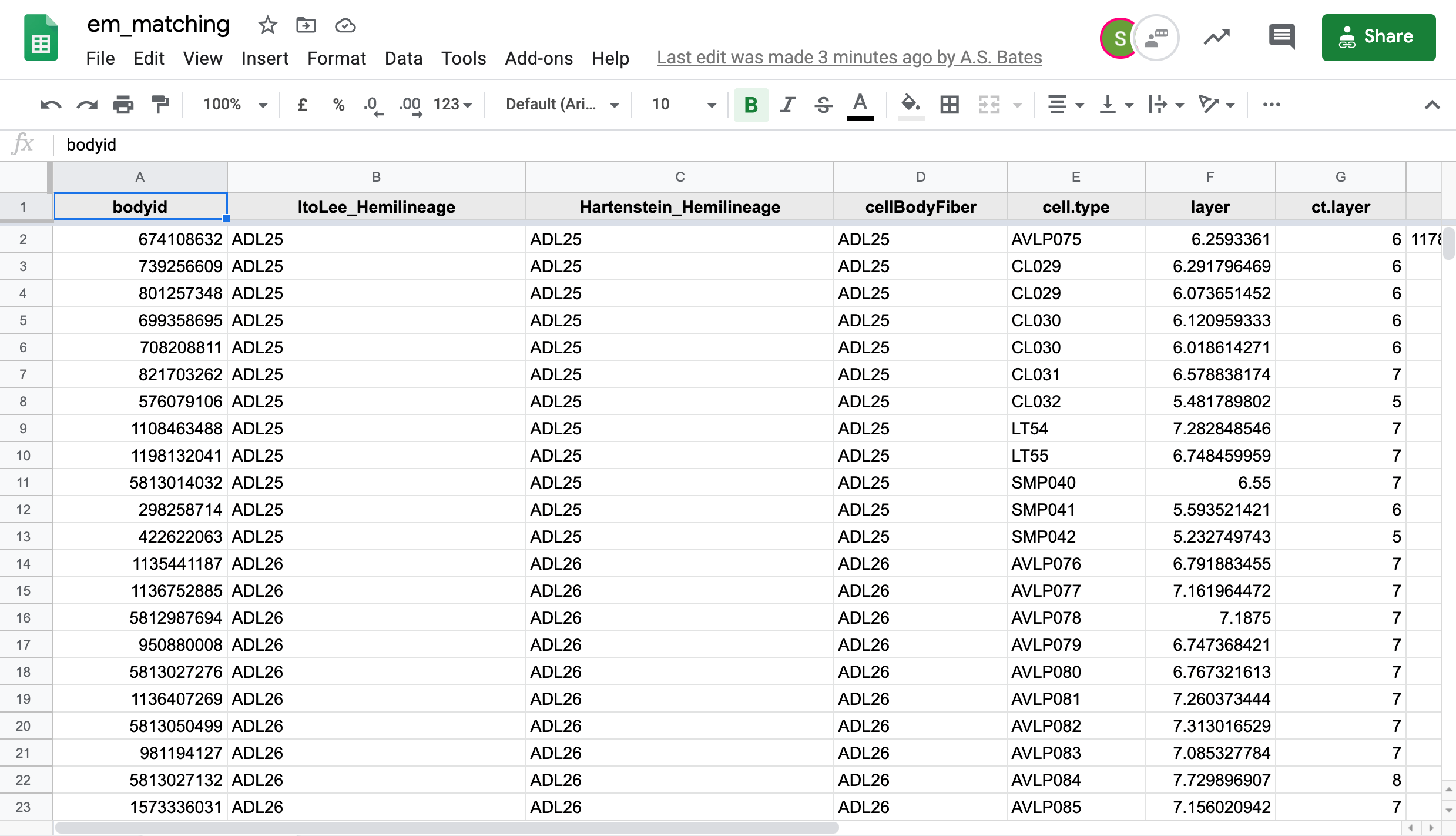 +
+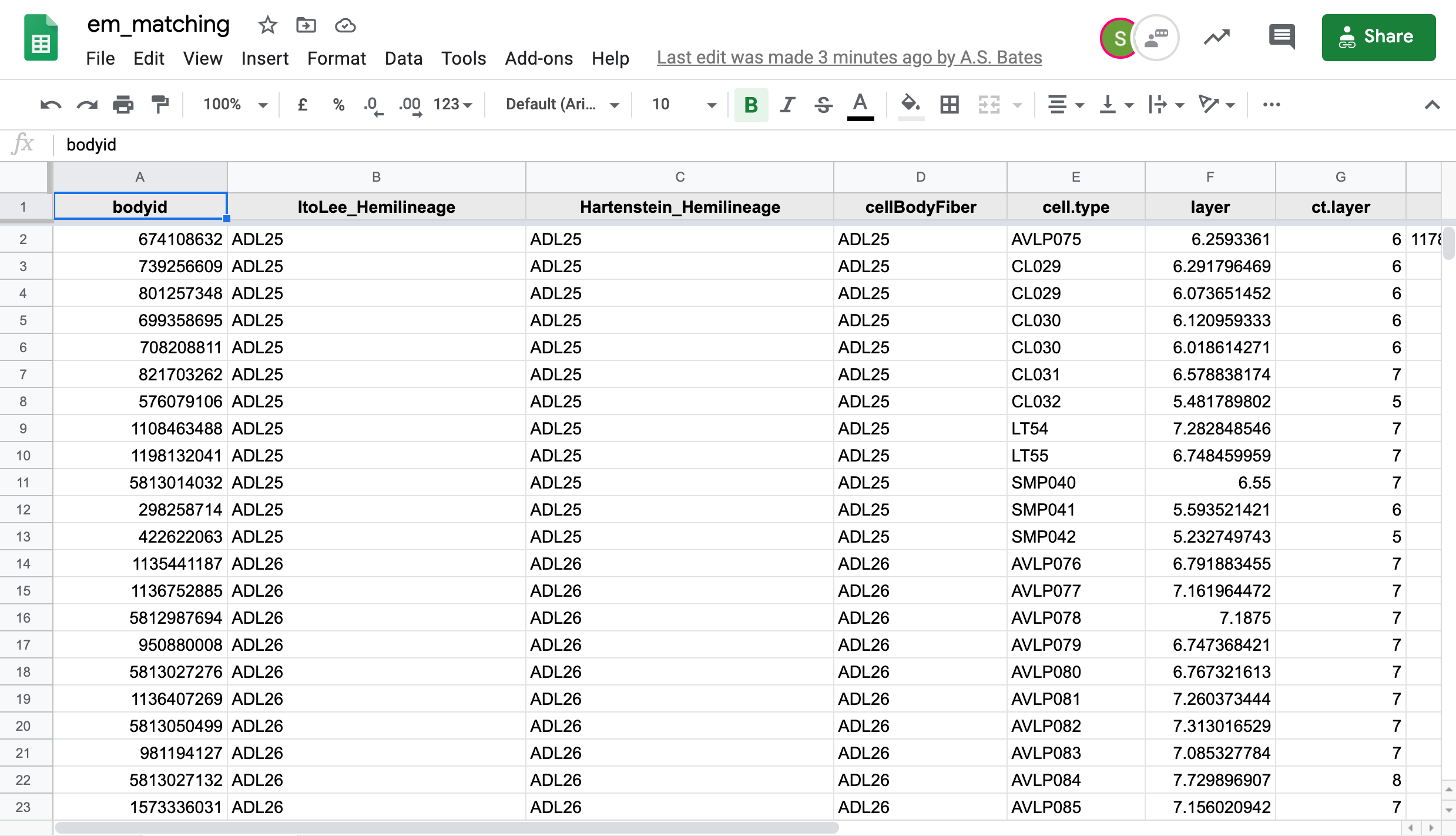
If you have authorisation, you can see the most up-to-date matches as so:
@@ -190,48 +193,48 @@
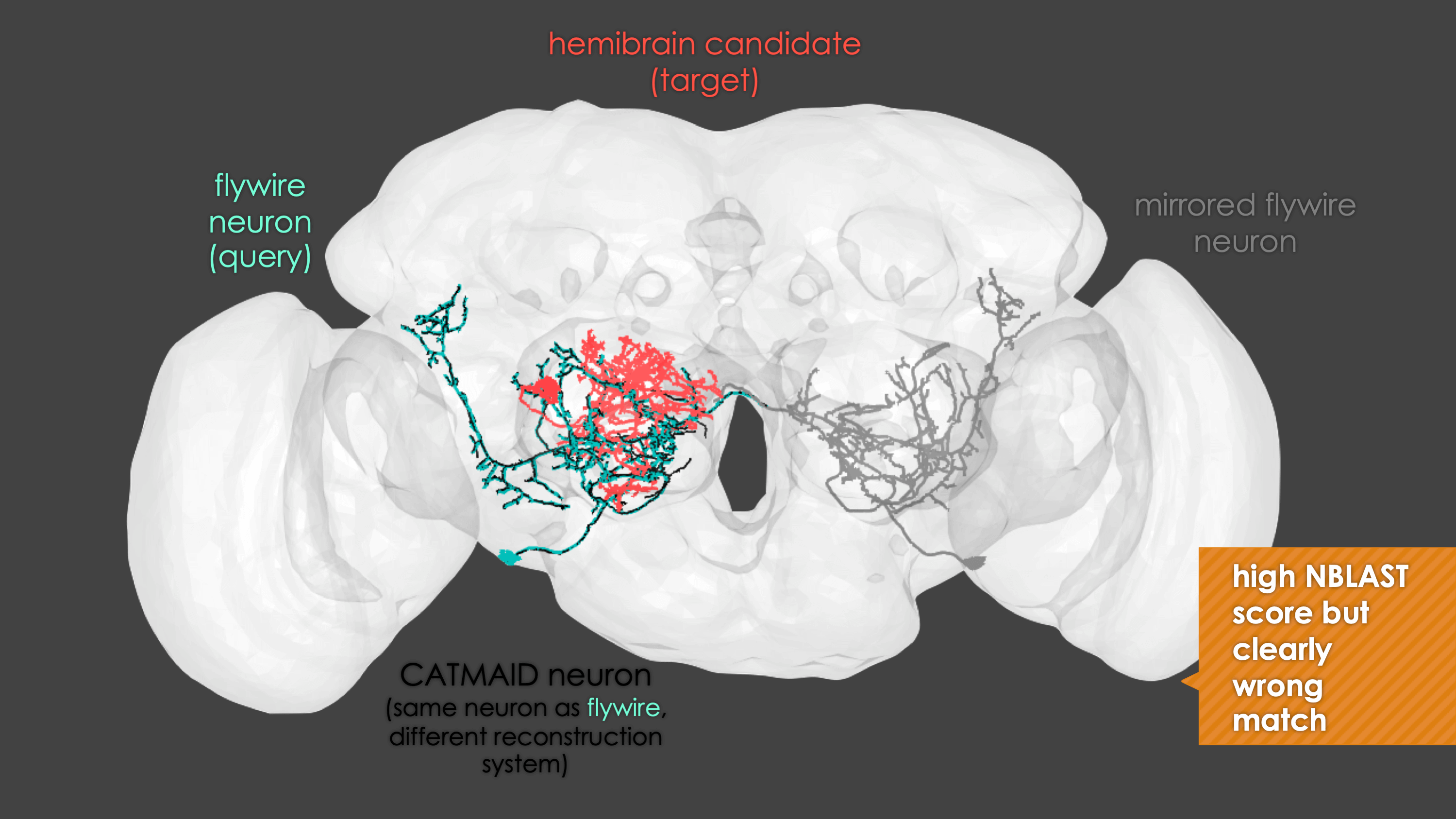
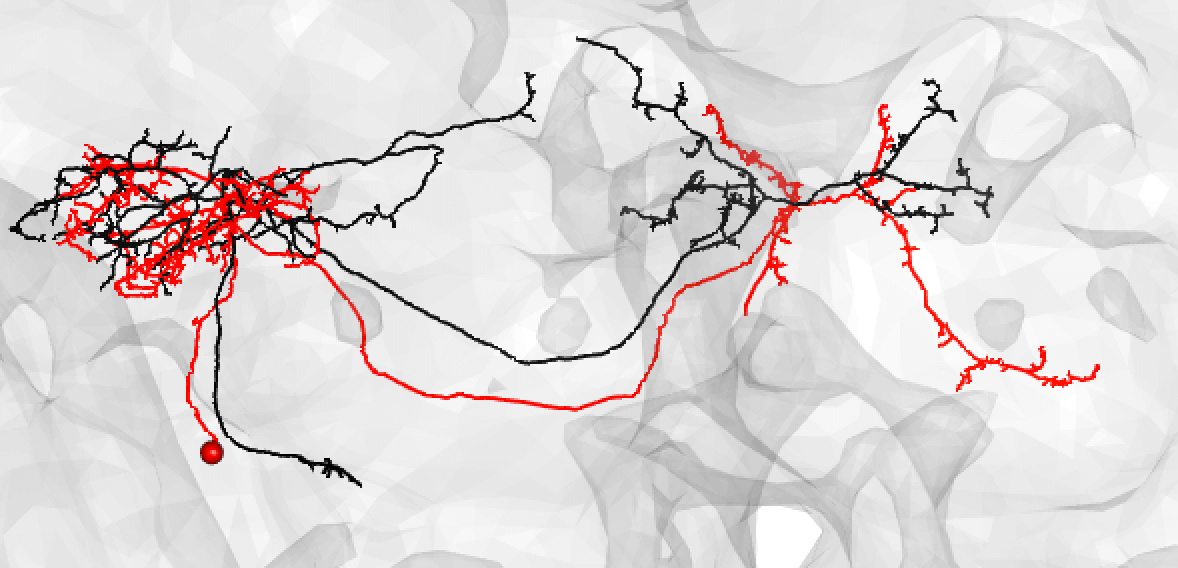
It is very important to note that a match cannot be a match if neurons do not seem to share the same cell body fiber tract. Being in a different tract is a deal breaker.
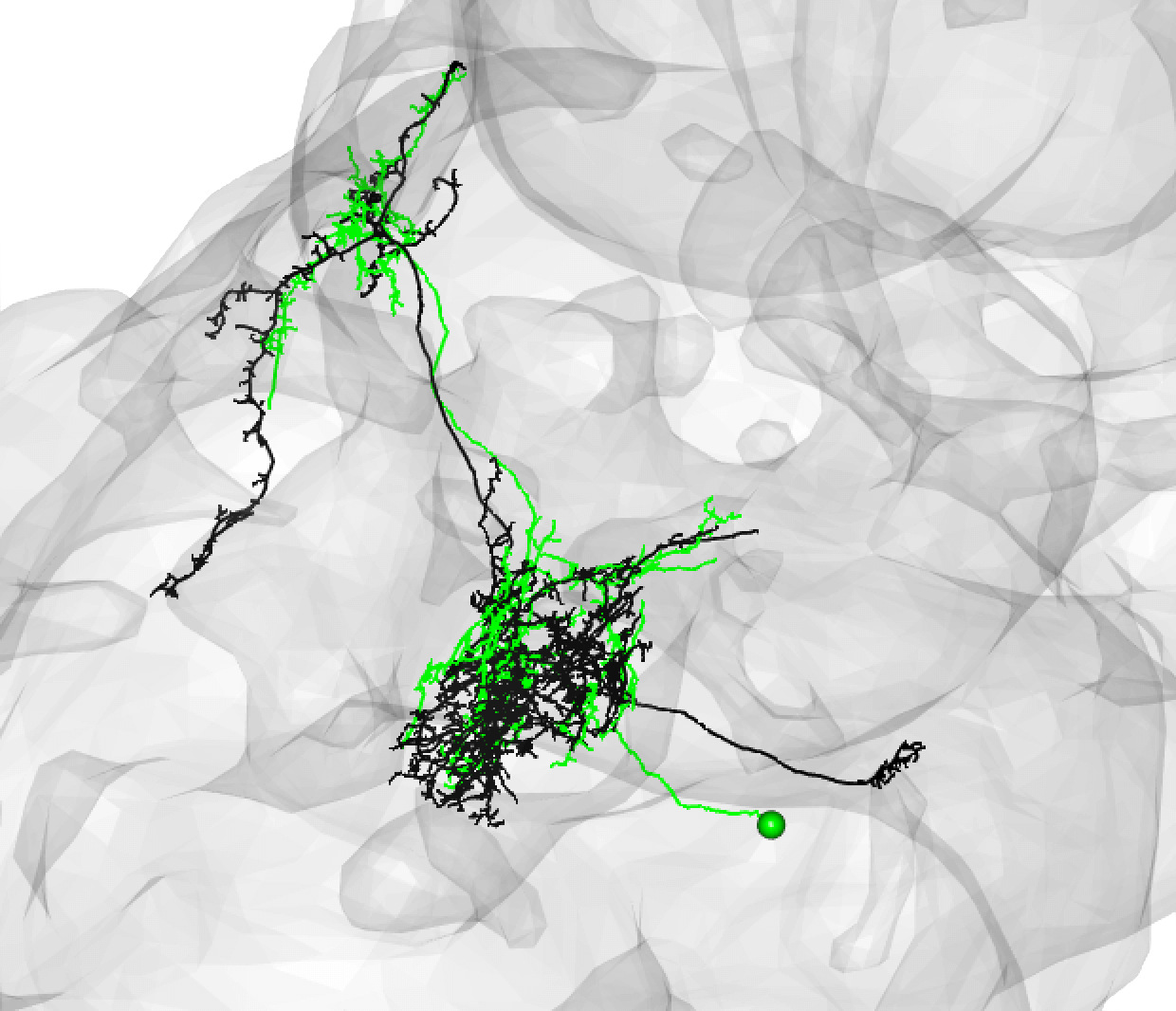
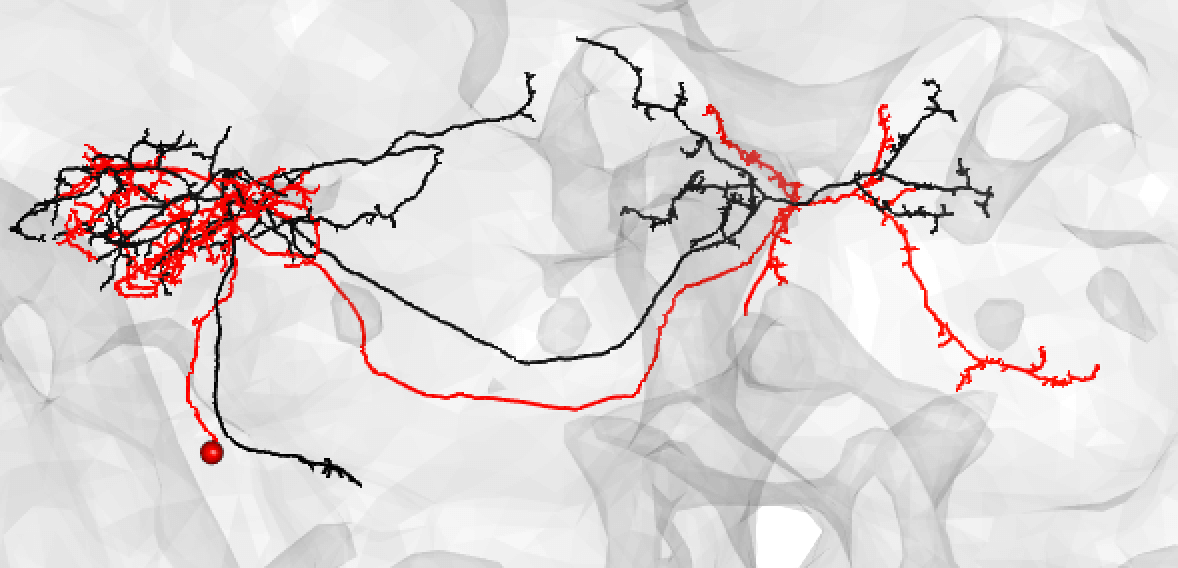
Some good matches are striking. For example:
- 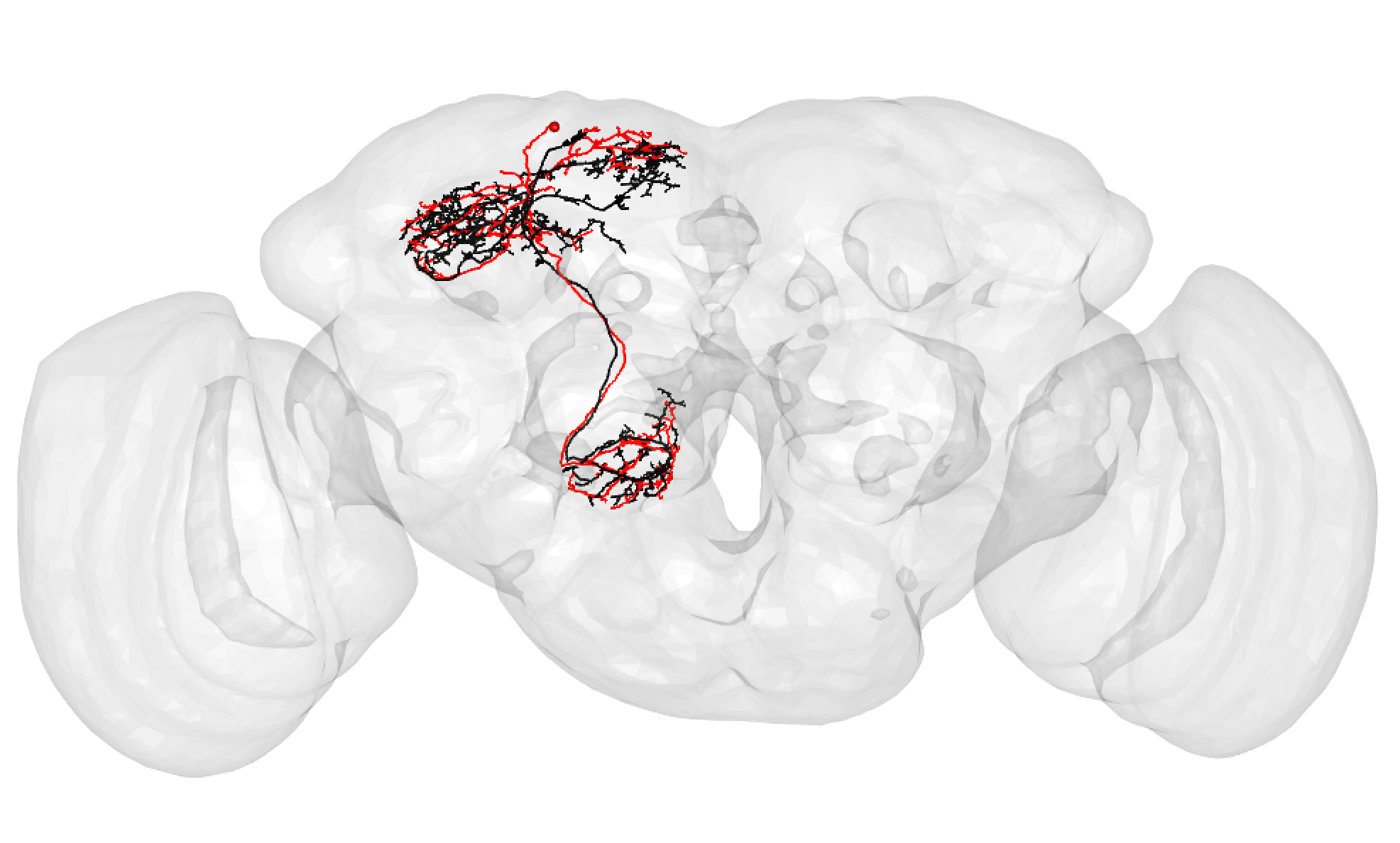 +
+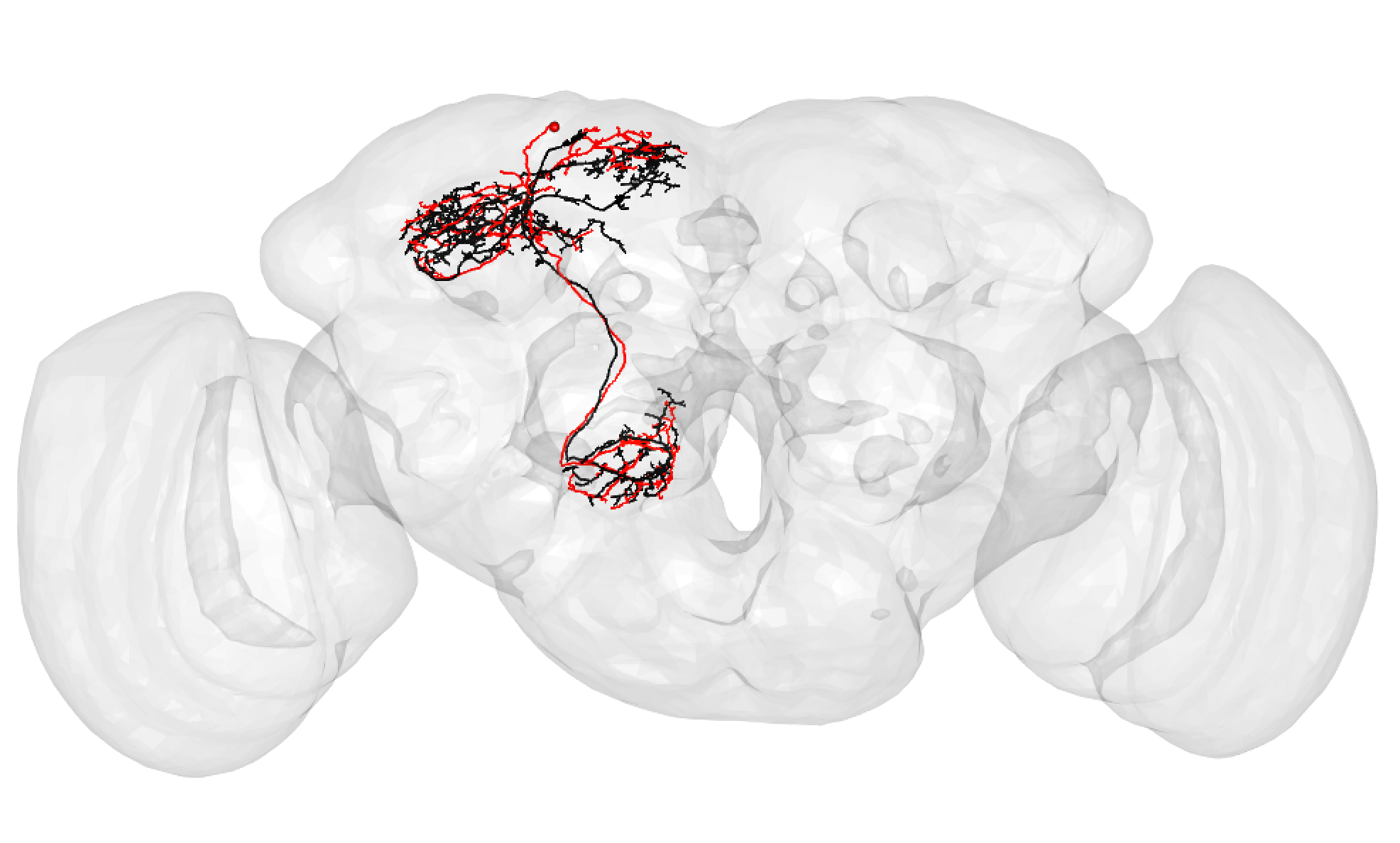
In the above case, the FAFB neuron has been quite extensively manually traced, meaning that these cells look very similar to one another.
Be aware that while neurons must share the same cell body fiber tract, these tracts can be a little off set. For example, this is also a good match:
- 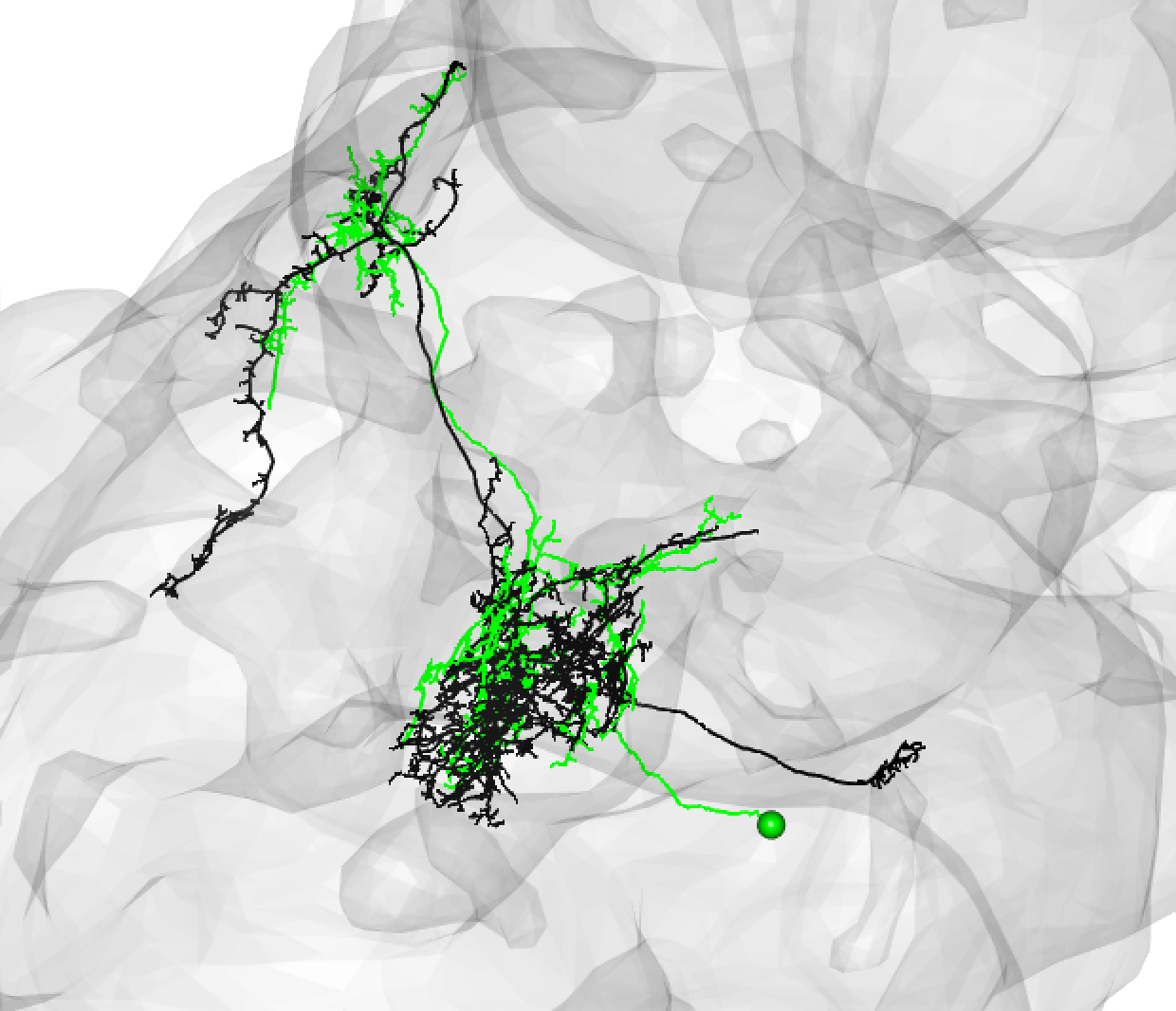 +
+
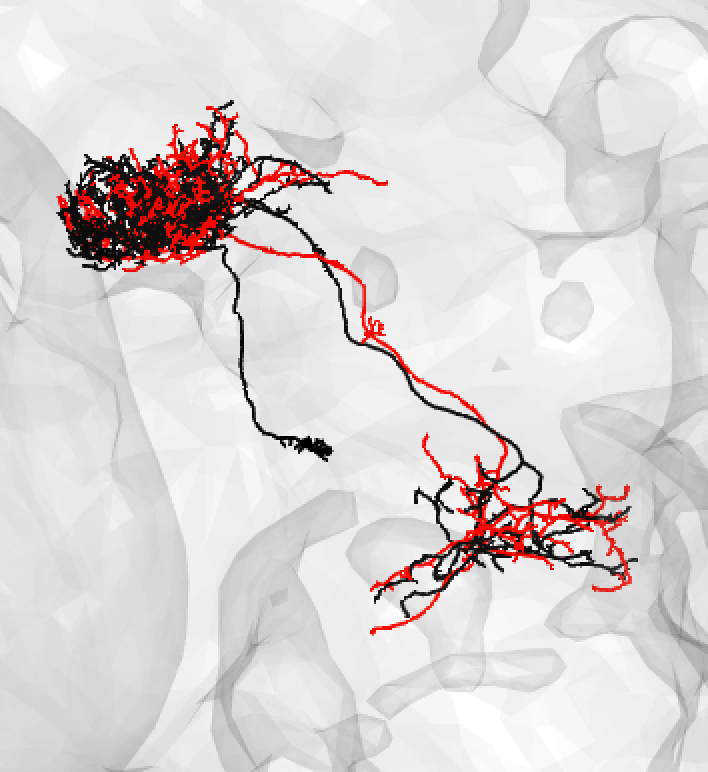
If the soma is missing, it might be safer to note a match as ‘medium’.
- 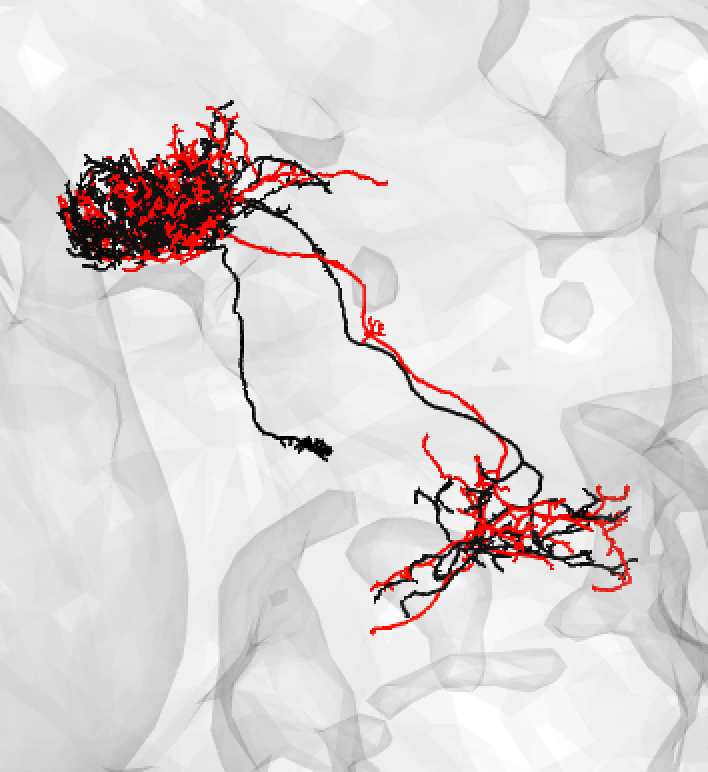 +
+
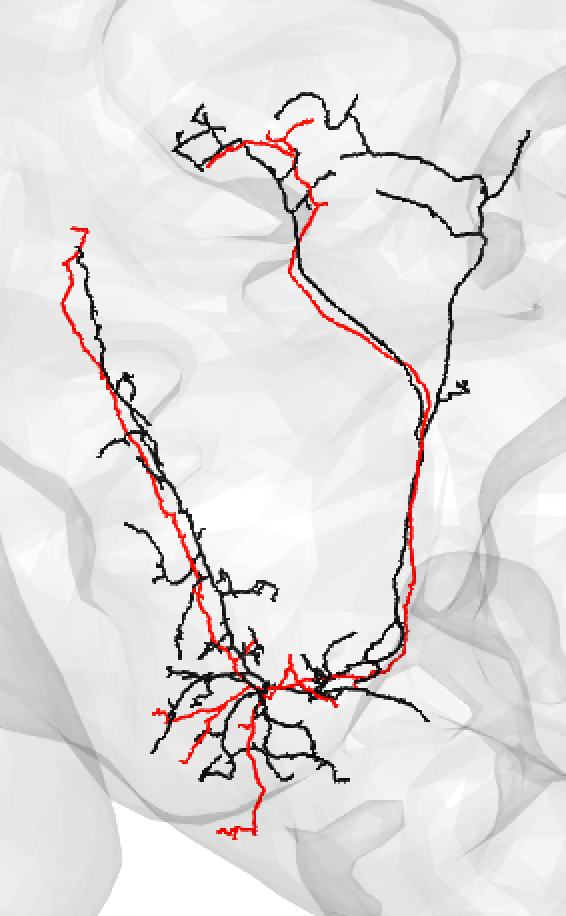
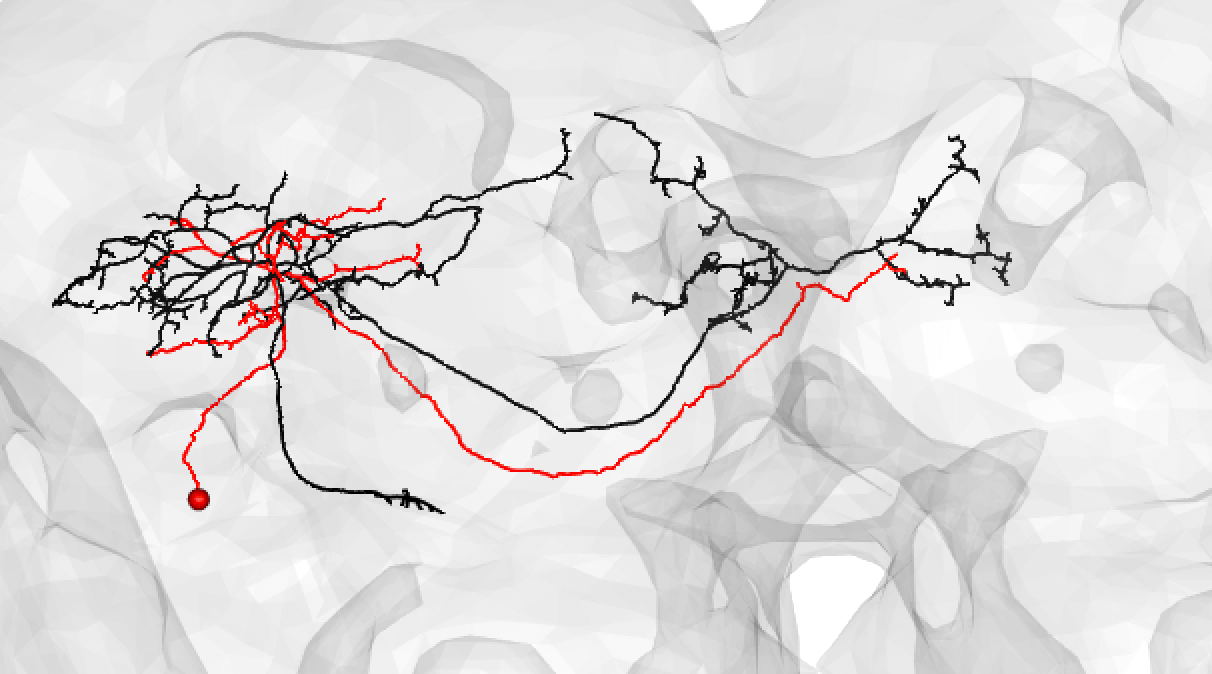
You might also use medium if you have a nice looking match and suspect that there is a medium/large discrepancy because the FAFB neuron (here shown in red) is under-traced, such as:
- 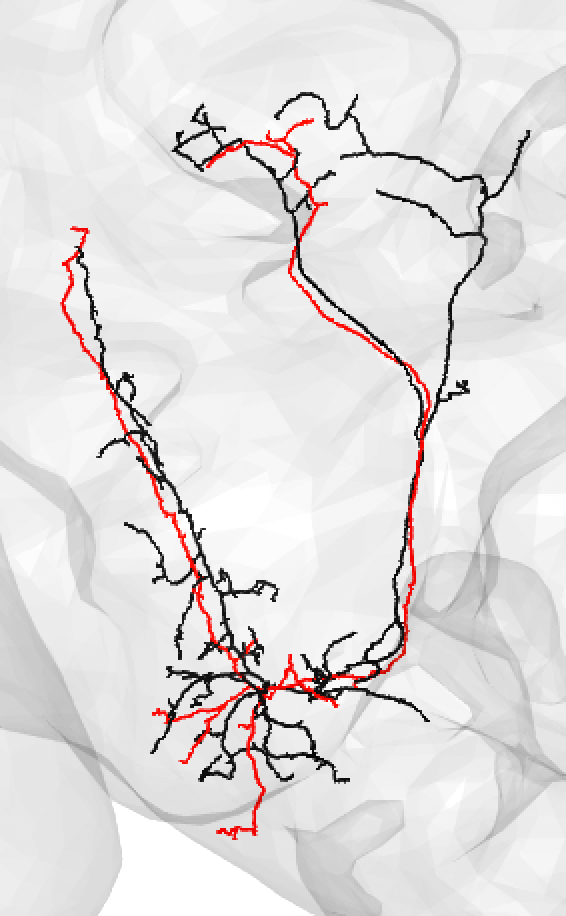 +
+
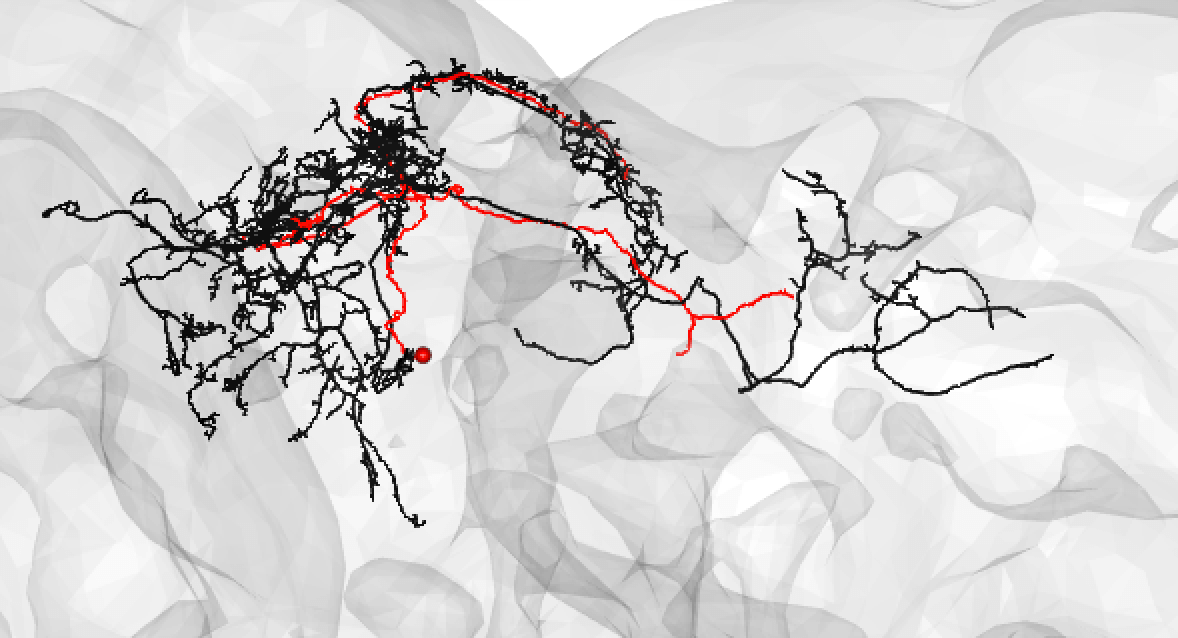
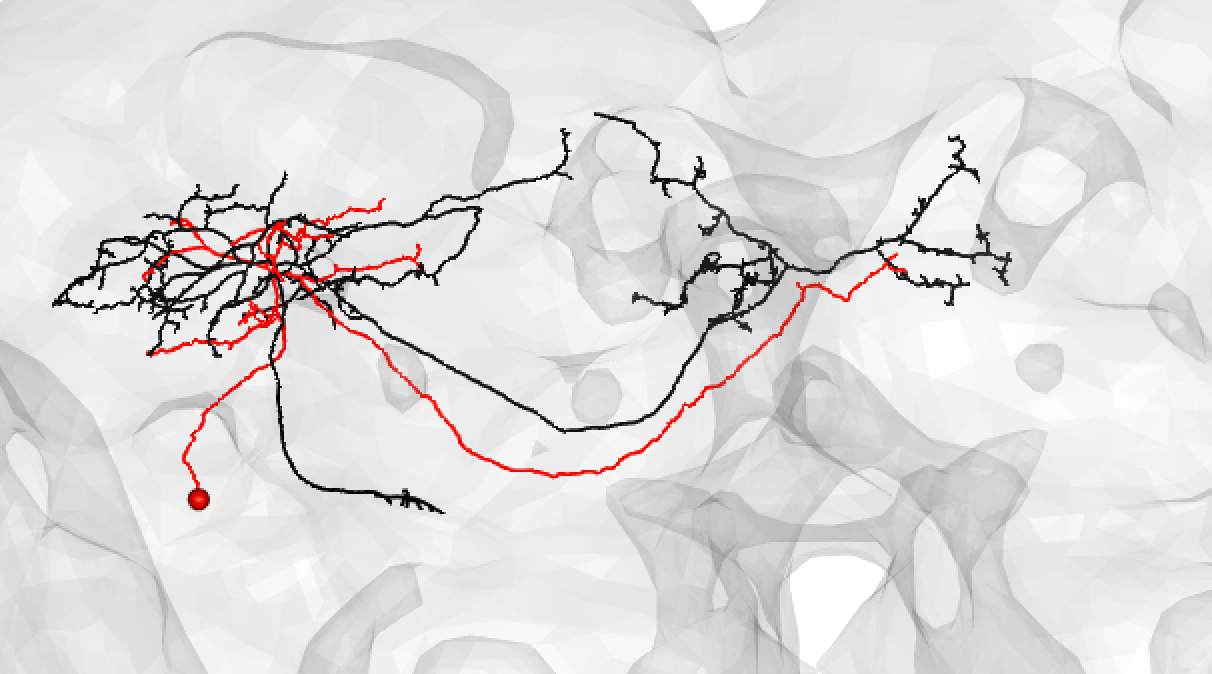
Or:
- 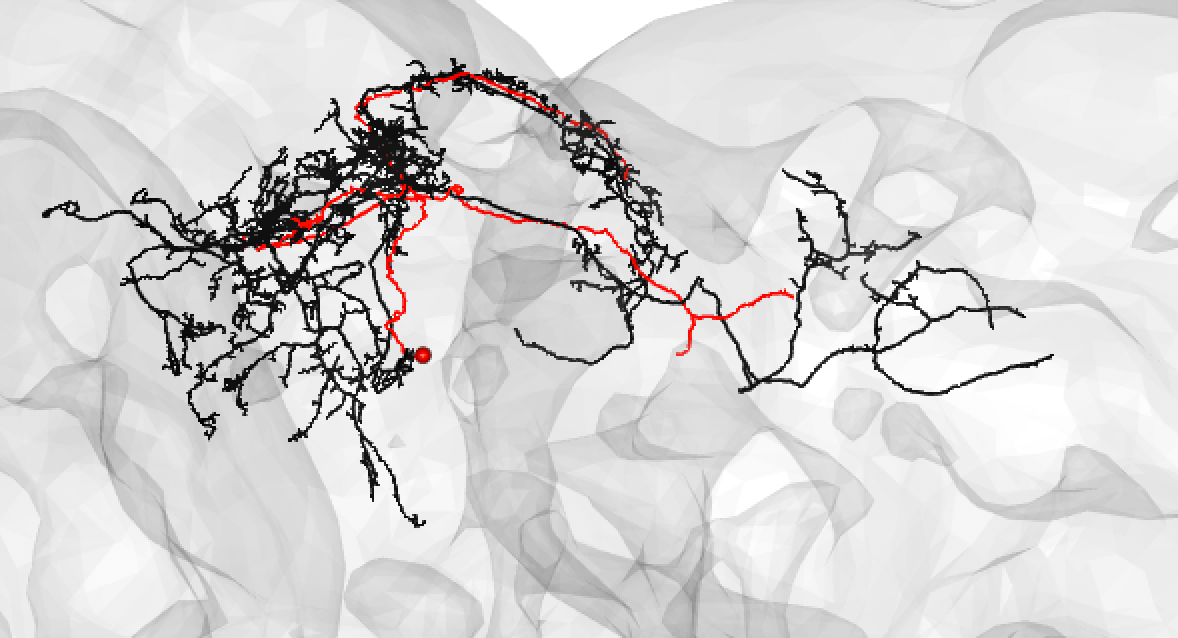 +
+
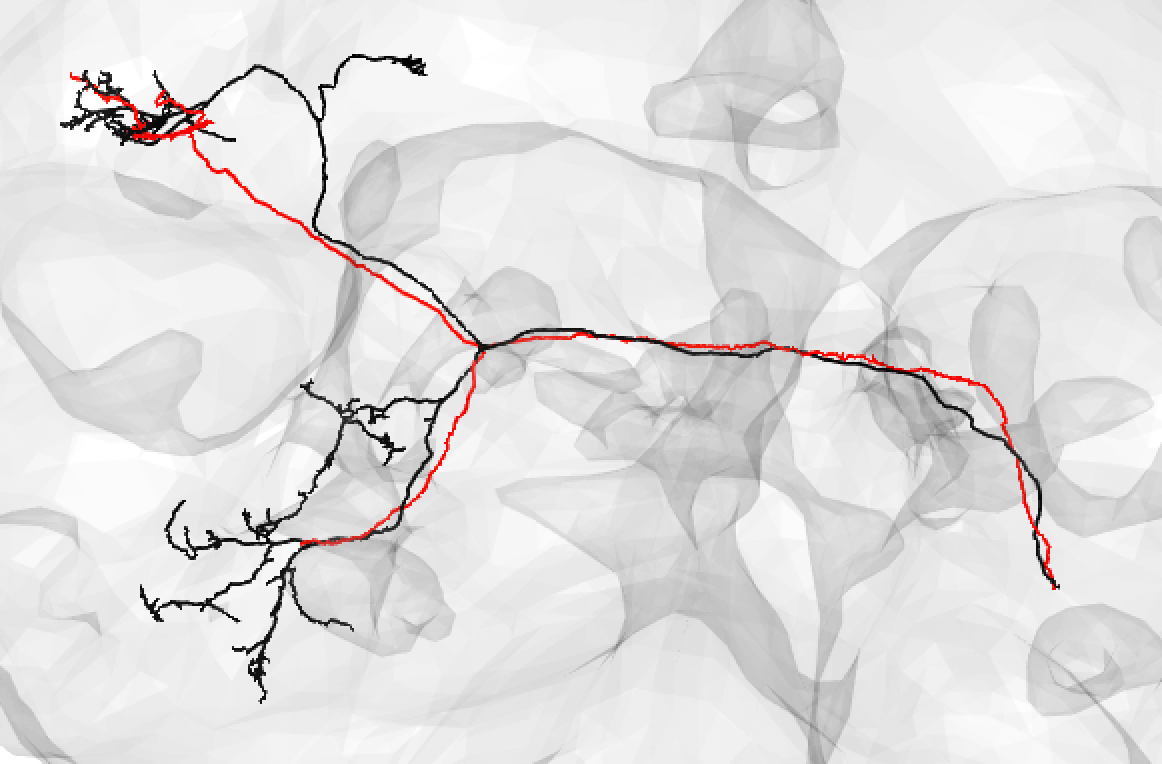
Bear in mind that the hemibrain volume only covers ~1/4 of the fly mid-brain, so neurons are truncated (here hemibrain neuron in black) but we can still make matches for many of them:
-  +
+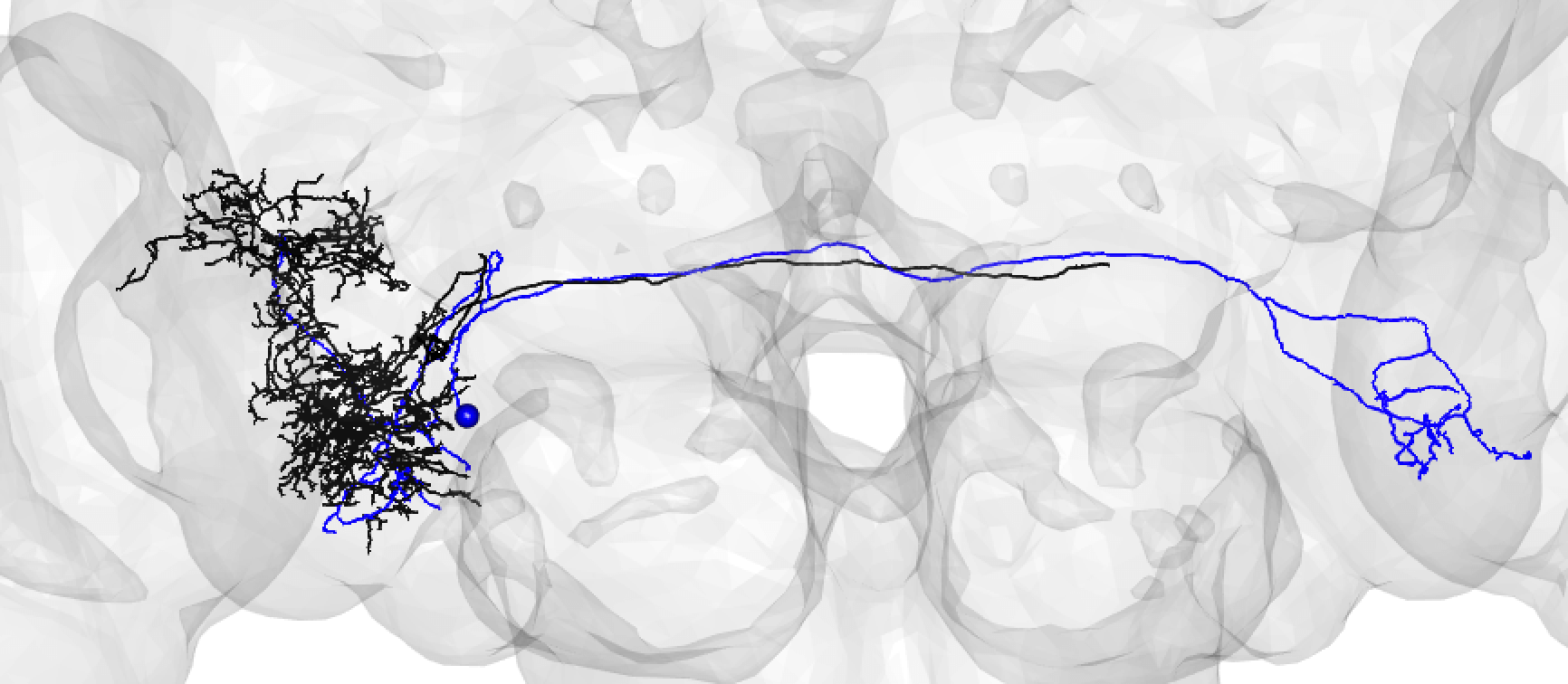
A larger degree of under-tracing may lead you to assign a match as poor. In this case, you think the two neurons may be ‘the same isomorphic cell type’ but you could be wrong. For example:
- 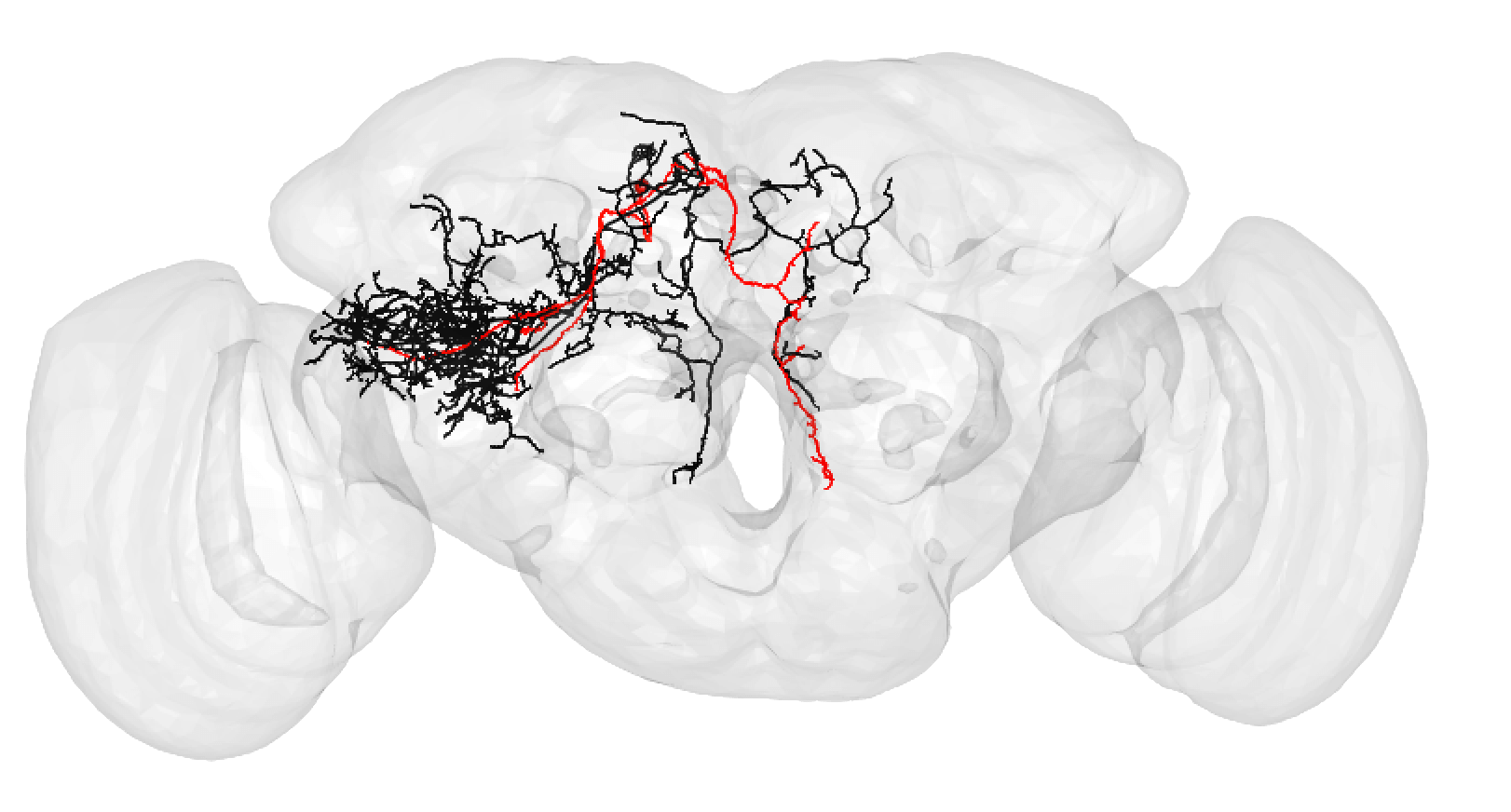 +
+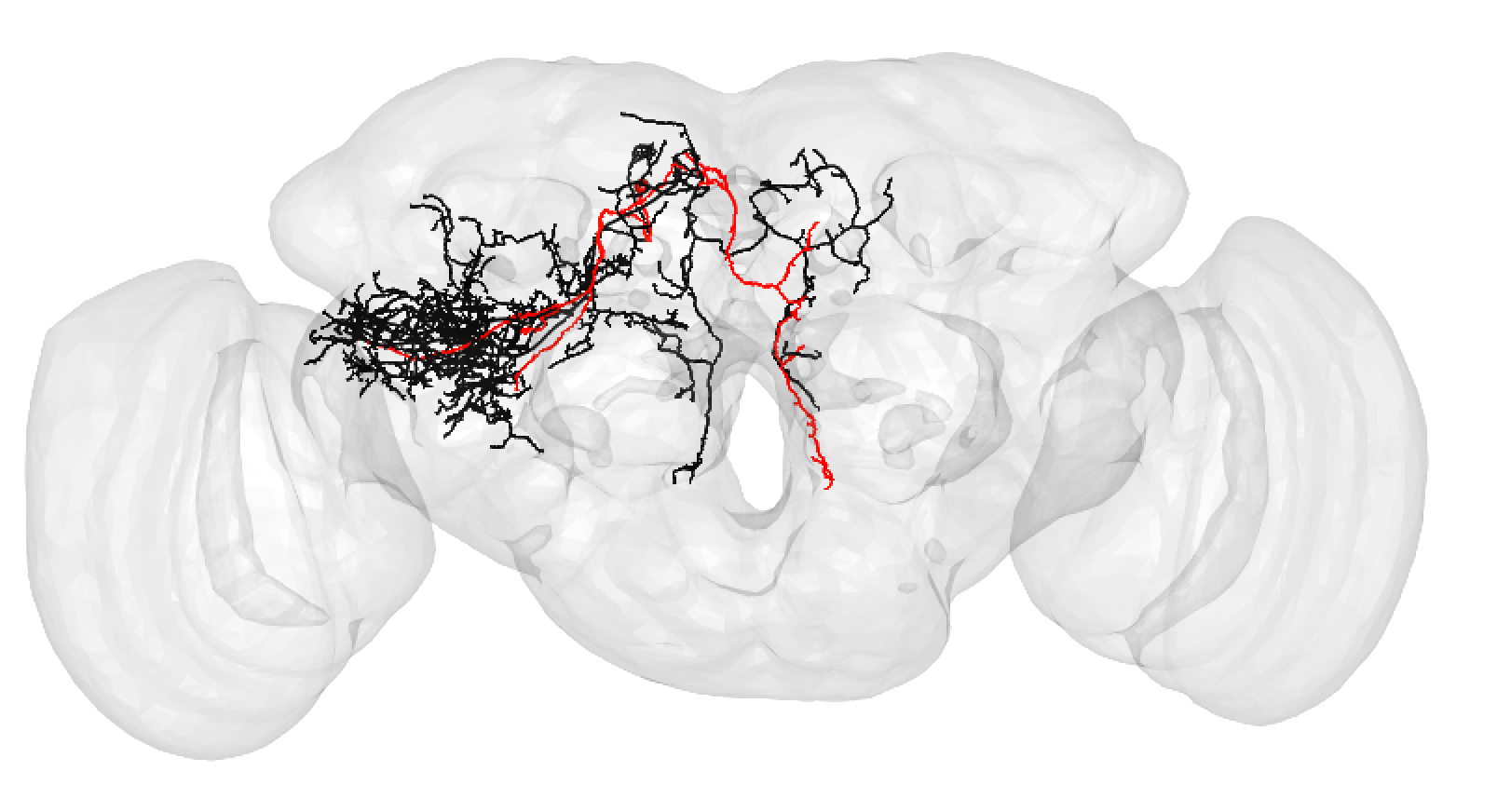
A poor match may also be made if you think there is a slight offset, possibly due to a registration issue:
-  +
+
Though in this case, choosing an even lesser-traced FAFB neuron may be better:
-  +
+
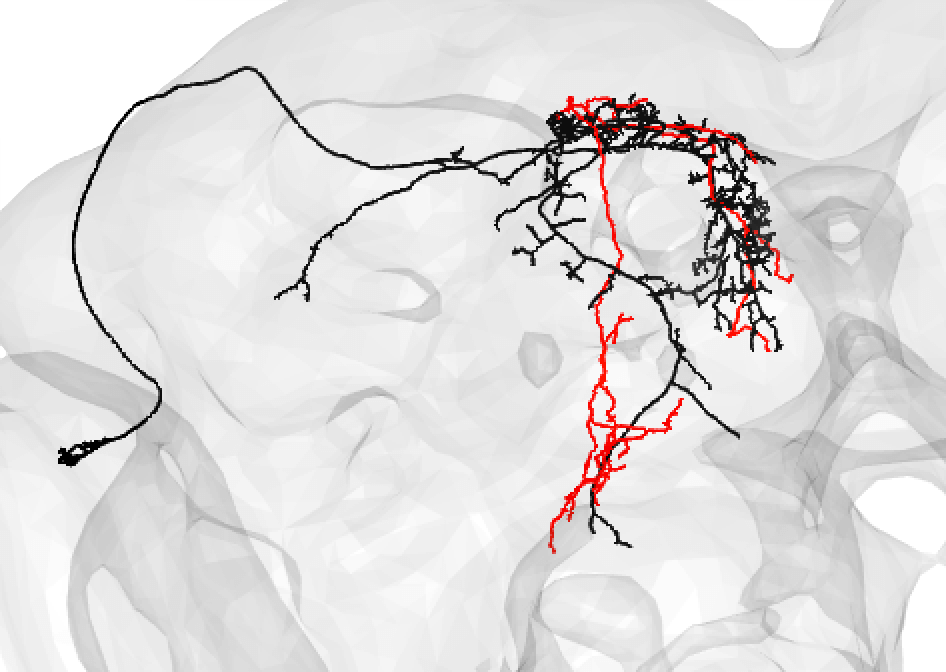
A poor match can be given even to very under-traced FAFB neurons:
-  +
+
And even fragments if you are convinced the morphology is unique enough (but be careful!):
- 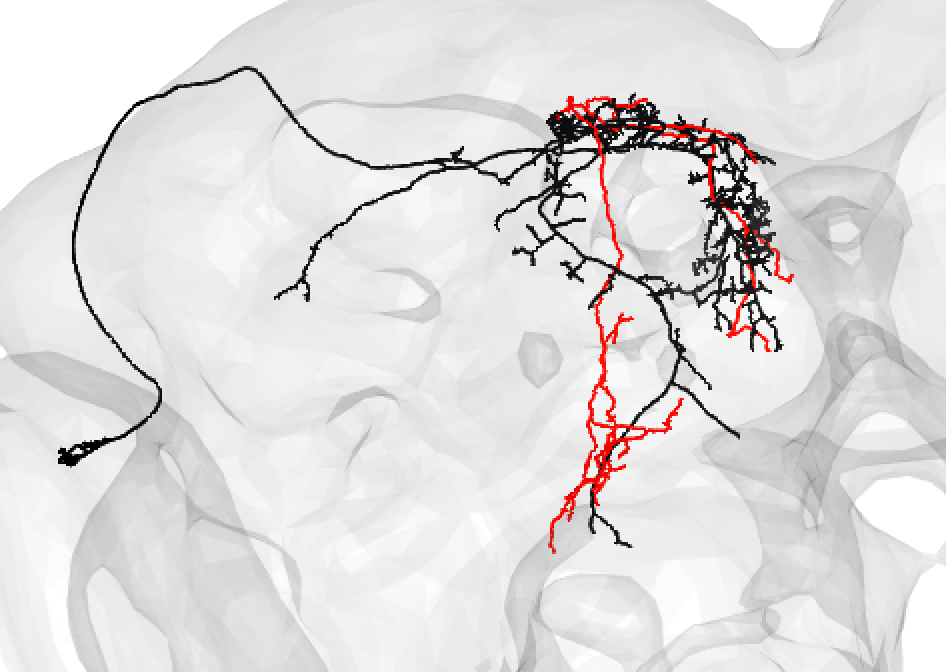 +
+
@@ -278,7 +281,7 @@
fafb_matching(ids = "16", overwrite = "none") # Re-look only if no proper match, or just a tract-only match, was found before.
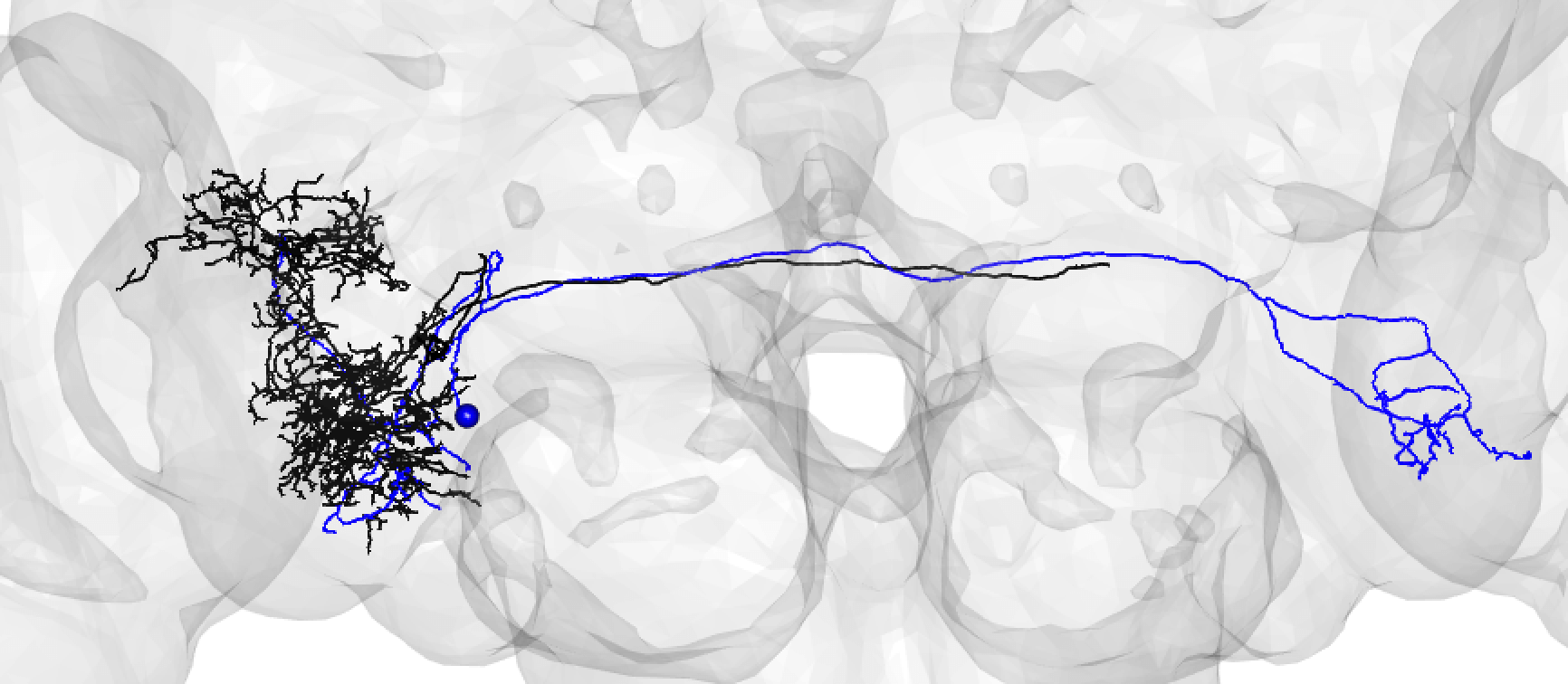
When you run these functions you will enter an interactive pipeline in an
rglwindow. Prompts will be given to you in your R console and you can rotate and pan in the window to see neurons. The neuron selected for-matching is shown in blue (i.e. if usinghemibrain_matchingthis will be a hemibrain neuron), and potential matches in red (i.e. if usinghemibrain_matchingthese will be FAFB neurons). Potential matches are shown by NBLAST score (a measure of morphological similarity). Usually, for reasonably traced FAFB neurons, a good match appears in the top 10 hits.-  +
+ @@ -306,7 +309,7 @@
@@ -306,7 +309,7 @@Uses
One use we have already found for all of this match making, is to cross-identify neuron cell body fiber tracts and (hemi)lineages. This means that we now have the locations in FAFB for different known sets of cells. You can see seed planes for them here.
-  +
+
- + hemibrain and flywire meta data + +
- flywire
-
@@ -138,7 +141,7 @@
hemibrainr
-The goal of hemibrainr is to provide useful code for preprocessing and analysing data from the Janelia FlyEM hemibrain project. It contains specific functionality for splitting neurons into axons and dendrites, including split edgelists for connectivity. It also has the capability to load large amount of precomputed data on flywire, FAFB and hemibrain neurons from a linked Google drive. Please see this article.
+The goal of hemibrainr is to provide useful code for preprocessing and analysing data from the Janelia FlyEM hemibrain project. It contains specific functionality for splitting neurons into axons and dendrites, including split edgelists for connectivity. It also has the capability to load large amount of precomputed data on flywire, FAFB and hemibrain neurons from a linked Google drive. Please see this article.
@@ -303,8 +306,8 @@@@ -146,7 +149,7 @@
navis
-navis (neuron analysis and visualization) is the Python equivalent of R’s nat and a lot of the other packages are built on top of it. It handles data representing neurons, such as skeletons or meshes, and lets you analyze and manipulate it.
+navishas a quickstart guide, an extensive API documentation and several [tutorials(https://navis.readthedocs.io/en/latest/source/gallery.html) including one to fetch hemibrain data via neuprint.navisalso provides an interfaces with R natverse functions for examplenat.nblastorxform_brain(see tutorials)navis (neuron analysis and visualization) is the Python equivalent of R’s nat and a lot of the other packages are built on top of it. It handles data representing neurons, such as skeletons or meshes, and lets you analyse and manipulate it.
navishas a quickstart guide, an extensive API documentation and several [tutorials(https://navis.readthedocs.io/en/latest/source/gallery.html) including one to fetch hemibrain data via neuprint.navisalso provides an interfaces with R natverse functions for examplenat.nblastorxform_brain(see tutorials)diff --git a/docs/authors.html b/docs/authors.html index 9f3a2e30..8ff2b467 100644 --- a/docs/authors.html +++ b/docs/authors.html @@ -95,6 +95,9 @@
-
+
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/favicon-16x16.png b/docs/favicon-16x16.png index 99b47eff..410b11fa 100644 Binary files a/docs/favicon-16x16.png and b/docs/favicon-16x16.png differ diff --git a/docs/favicon-32x32.png b/docs/favicon-32x32.png index d4f44fea..84426d89 100644 Binary files a/docs/favicon-32x32.png and b/docs/favicon-32x32.png differ diff --git a/docs/index.html b/docs/index.html index eb1d4736..aef9c186 100644 --- a/docs/index.html +++ b/docs/index.html @@ -56,6 +56,9 @@
- + hemibrain and flywire meta data + +
- flywire
-
@@ -106,7 +109,7 @@
The goal of hemibrainr is to provide useful code for preprocessing and analysing data from the Janelia FlyEM hemibrain project. It makes use of the natverse R package, neuprintr to get hemibrain data from their connectome analysis and data hosting service neuprint. The dataset has been described here. Using this R package in concert with the natverse ecosystem is highly recommended.
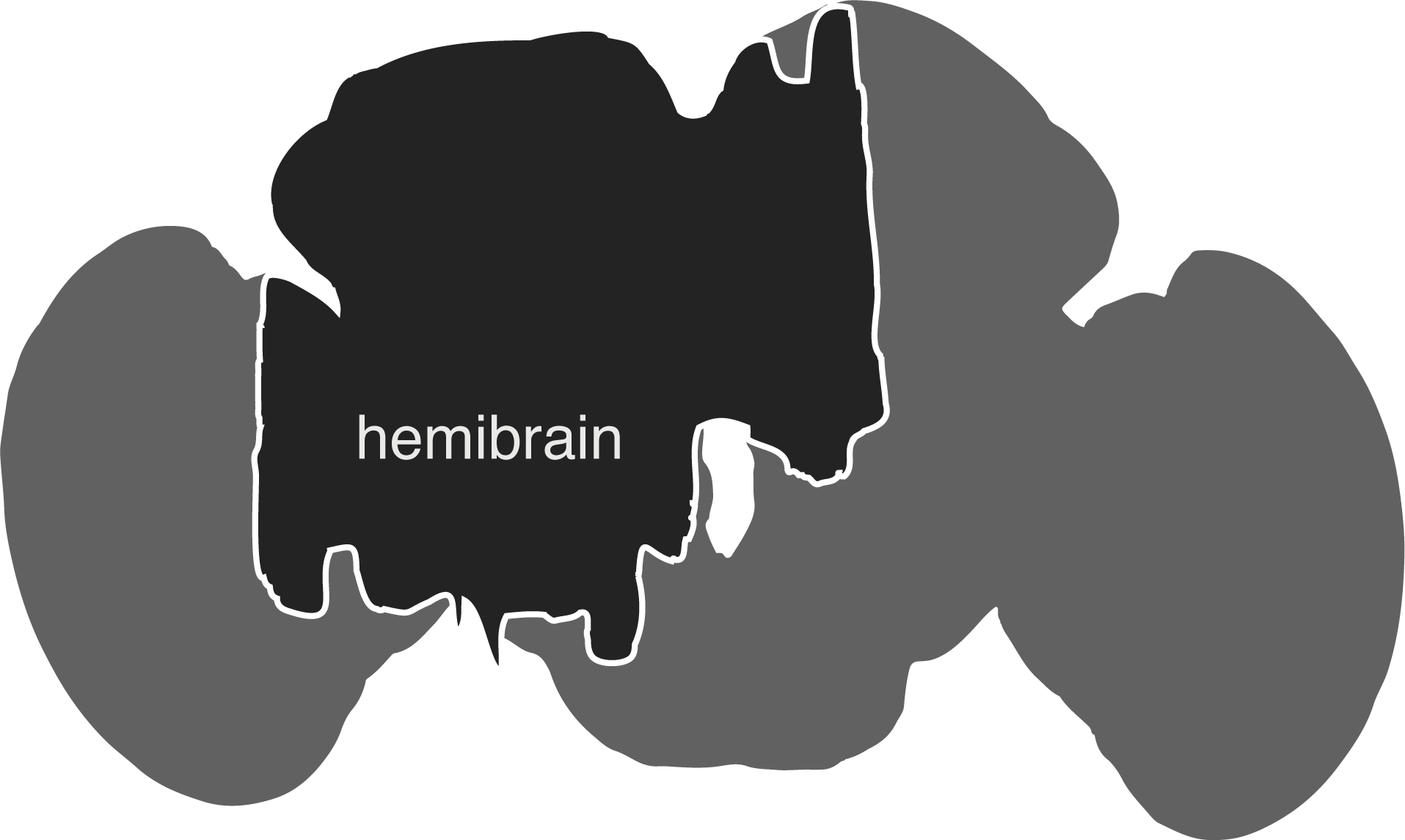
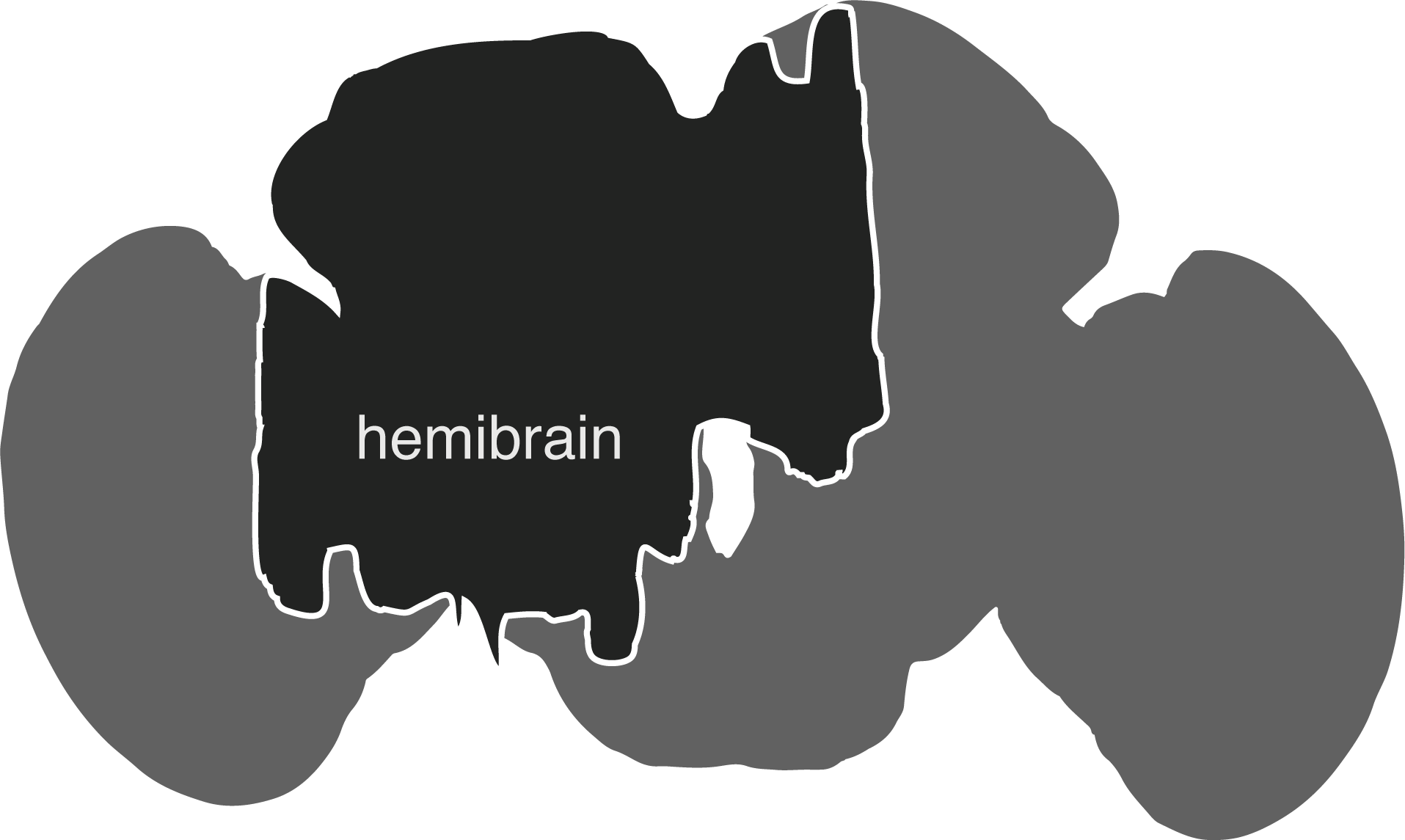
The hemibrain connectome comprises the region of the fly brain depicted below. It is ~21,662 ~full neurons, 9.5 million synapses and is about ~35% complete in this region:
-
+
 -
-@@ -117,7 +120,7 @@
@@ -184,7 +187,7 @@# install if (!require("remotes")) install.packages("remotes") -remotes::install_github("flyconnectome/hemibrainr") +remotes::install_github("natverse/hemibrainr") # use library(hemibrainr)For this, you need access to th hemibrainr google team drive. Authentication is through an email account. Once you have access, there are two basic ways to mount the data for use:
Option 1, mount your Google drives using Google filestream. However, for this to work you will need Google Workspace, Google’s monthly subscription offering for businesses and organizations. One the Google filestream application is run, you should be able to see your drives mounted like external hard drive, as so:
-  +
+
Then, this should work:
-@@ -194,7 +197,7 @@
# Now just get the name of your default team drive. ## This will be used to locate your team drive using the R package googledrive hemibrainr_team_drive()
Option 2, this is free. You still need authenticated access to the hemibrainr Gogle team drive. It cna then be moutned using rclone. First, download rclone for your operating system. You can also download from your system’s command line (e.g. from terminal) and then configure it for the drive:
+Option 2, this is free. You still need authenticated access to the hemibrainr Gogle team drive. It can then be mounted using rclone. First, download rclone for your operating system. You can also download from your system’s command line (e.g. from terminal) and then configure it for the drive:
@@ -216,7 +219,7 @@# And now we are back to: options("Gdrive_hemibrain_data")
For more detailed instructions, see this article.
+For more detailed instructions, see this article.
@@ -270,7 +273,7 @@
This package was created by Alexander Shakeel Bates and Gregory Jefferis. You can cite this package as:
-citation(package = "hemibrainr")Bates AS, Jefferis GSXE (2020). hemibrainr: Code for working with data from Janelia FlyEM’s hemibrain project. R package version 0.1.0. https://github.com/flyconnectome/hemibrainr
+Bates AS, Jefferis GSXE (2020). hemibrainr: Code for working with data from Janelia FlyEM’s hemibrain project. R package version 0.1.0. https://github.com/natverse/hemibrainr
Developers
diff --git a/docs/pkgdown.yml b/docs/pkgdown.yml index ca80c1d6..d7b4900f 100644 --- a/docs/pkgdown.yml +++ b/docs/pkgdown.yml @@ -2,6 +2,7 @@ pandoc: 2.3.1 pkgdown: 1.6.1.9000 pkgdown_sha: 6094ac3dd80a1f3f1d4e2f47d0ed4716d51362b0 articles: + data: data.html flywire: flywire.html google_filestream: google_filestream.html hemibrain_alpns_toons: hemibrain_alpns_toons.html @@ -9,5 +10,5 @@ articles: hemibrain_connectivity: hemibrain_connectivity.html match_making: match_making.html packages_guide: packages_guide.html -last_built: 2020-12-04T19:45Z +last_built: 2021-01-06T17:54Z diff --git a/docs/reference/add_Label.html b/docs/reference/add_Label.html index 7b210334..e83b2195 100644 --- a/docs/reference/add_Label.html +++ b/docs/reference/add_Label.html @@ -97,6 +97,9 @@-
+
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/add_field.html b/docs/reference/add_field.html index df2c4f52..c74a4ee7 100644 --- a/docs/reference/add_field.html +++ b/docs/reference/add_field.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/class2ids.html b/docs/reference/class2ids.html index 27f6b247..92522c40 100644 --- a/docs/reference/class2ids.html +++ b/docs/reference/class2ids.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/classed.ids.html b/docs/reference/classed.ids.html index 17386dbe..d726c7d7 100644 --- a/docs/reference/classed.ids.html +++ b/docs/reference/classed.ids.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -185,8 +188,6 @@
-
+
-
+
-
+
Bodyids for neuron classes
lc.ids -kc.ids - rn.info
All vignettes
-
+
FormatAn object of class
characterof length 307.An object of class
characterof length 2383.An object of class
-characterof length 1428.An object of class
+characterof length 431.An object of class
characterof length 345.An object of class
characterof length 1927.An object of class
characterof length 5.An object of class
characterof length 24.An object of class
-characterof length 2129.An object of class
characterof length 1927.An object of class
data.framewith 2651 rows and 44 columns.Details
diff --git a/docs/reference/extract_cable.html b/docs/reference/extract_cable.html index b122bb0e..d1721dbc 100644 --- a/docs/reference/extract_cable.html +++ b/docs/reference/extract_cable.html @@ -96,6 +96,9 @@-
+
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/fafb_hemibrain_annotate.html b/docs/reference/fafb_hemibrain_annotate.html index 416296f2..cfe8c579 100644 --- a/docs/reference/fafb_hemibrain_annotate.html +++ b/docs/reference/fafb_hemibrain_annotate.html @@ -49,7 +49,7 @@ +Not that catmaid::flywire_matching_rewrite() will write annotations related to flywire." /> @@ -98,6 +98,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -150,7 +153,7 @@
-
+
Set annotations for FAFB neurons in CATMAID based on hemibrain results
Set annotations for FAFB neurons in CATMAID based on matches made to hemibrain neurons. E.g. transfer information on matches and cell body fibers, and also update lineage related information. -Not that
+Not thatcatmaid::flywire_matching_rewrite()will writ annotations related to flywire.catmaid::flywire_matching_rewrite()will write annotations related to flywire.fafb_hemibrain_annotate(x, flywire = TRUE, ...) diff --git a/docs/reference/flow_centrality.html b/docs/reference/flow_centrality.html index d31da248..b0cfd07d 100644 --- a/docs/reference/flow_centrality.html +++ b/docs/reference/flow_centrality.html @@ -100,6 +100,9 @@
-
+
- + hemibrain and flywire meta data +
- flywire @@ -281,18 +284,18 @@
Examp # Get neurons neurons = neuprintr::neuprint_read_neurons(tough) - +
#> Error in neuprintr::neuprint_read_neurons(tough): Error reading bodyids# Now make sure the neurons have a soma marked ## Some hemibrain neurons do not, as the soma was chopped off neurons.checked = hemibrain_skeleton_check(neurons, meshes = hemibrain.surf) -#> Re-rooting 2 neurons without a soma#> Removing synapses outside the hemibrain neuropil volume for 5 neurons#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated+#> Error in nat::as.neuronlist(x): object 'neurons' not found# Split neuron ## These are the recommended parameters for hemibrain neurons neurons.flow = flow_centrality(neurons.checked, polypre = TRUE, mode = "centrifugal", split = "distance") - +#> Error in flow_centrality(neurons.checked, polypre = TRUE, mode = "centrifugal", split = "distance"): object 'neurons.checked' not foundif (FALSE) { # Plot the split to check it nat::nopen3d() diff --git a/docs/reference/flycircuit_neurons.html b/docs/reference/flycircuit_neurons.html index 08781a2e..c1505f02 100644 --- a/docs/reference/flycircuit_neurons.html +++ b/docs/reference/flycircuit_neurons.html @@ -98,6 +98,9 @@--
+
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/flywire_googledrive_data.html b/docs/reference/flywire_googledrive_data.html index c1b84279..3ff53d95 100644 --- a/docs/reference/flywire_googledrive_data.html +++ b/docs/reference/flywire_googledrive_data.html @@ -101,6 +101,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -159,7 +162,9 @@
-
+
Read precomputed flywire data from the hemibrainr Google Drive
scores retrieved usinghemibrain_nblast.flywire_meta(local = FALSE, folder = "flywire_neurons/", sql = TRUE, ...) +
flywire_meta(local = FALSE, folder = "flywire_neurons/", sql = FALSE, ...) + +flywire_failed(local = FALSE, folder = "flywire_neurons/", sql = FALSE, ...) flywire_contributions( local = FALSE, @@ -168,7 +173,7 @@
+flywire_ids(local = FALSE, folder = "flywire_neurons/", sql = FALSE, ...)Read precomputed flywire data from the hemibrainr Google Drive
... ) -flywire_ids(local = FALSE, folder = "flywire_neurons/", sql = TRUE, ...)Arguments
diff --git a/docs/reference/flywire_ids_update.html b/docs/reference/flywire_ids_update.html index c2aa7e04..212d3b4d 100644 --- a/docs/reference/flywire_ids_update.html +++ b/docs/reference/flywire_ids_update.html @@ -47,7 +47,7 @@ - @@ -99,6 +99,9 @@
diff --git a/docs/reference/paper_colours.html b/docs/reference/paper_colours.html index cb9380ac..e0818b69 100644 --- a/docs/reference/paper_colours.html +++ b/docs/reference/paper_colours.html @@ -96,6 +96,9 @@-
+
- + hemibrain and flywire meta data +
- flywire @@ -149,7 +152,7 @@
- + hemibrain and flywire meta data +
- flywire @@ -156,7 +160,8 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/grouped_adjacency.html b/docs/reference/grouped_adjacency.html index f4ed221a..b9d196b2 100644 --- a/docs/reference/grouped_adjacency.html +++ b/docs/reference/grouped_adjacency.html @@ -99,6 +99,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain.surf.html b/docs/reference/hemibrain.surf.html index a3bd2089..413d0763 100644 --- a/docs/reference/hemibrain.surf.html +++ b/docs/reference/hemibrain.surf.html @@ -99,6 +99,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_add_made_matches.html b/docs/reference/hemibrain_add_made_matches.html index 6ef27b9f..07728ee2 100644 --- a/docs/reference/hemibrain_add_made_matches.html +++ b/docs/reference/hemibrain_add_made_matches.html @@ -103,6 +103,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -166,7 +169,11 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_adjust_saved_split.html b/docs/reference/hemibrain_adjust_saved_split.html index 6dad15d4..e1d5baa0 100644 --- a/docs/reference/hemibrain_adjust_saved_split.html +++ b/docs/reference/hemibrain_adjust_saved_split.html @@ -102,6 +102,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_al.surf.html b/docs/reference/hemibrain_al.surf.html index 2b94f3fb..fe77dff1 100644 --- a/docs/reference/hemibrain_al.surf.html +++ b/docs/reference/hemibrain_al.surf.html @@ -101,6 +101,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -179,7 +182,7 @@
"excpected_cit"a citation that describes this glomerulus,
+"expected_cit"a citation that describes this glomerulus,
"expected_RN_female_1h".
"expected_RN_female_SD".
"missing".
@@ -195,7 +198,7 @@ - + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_clean_skeleton.html b/docs/reference/hemibrain_clean_skeleton.html index 58dddc1d..11b42939 100644 --- a/docs/reference/hemibrain_clean_skeleton.html +++ b/docs/reference/hemibrain_clean_skeleton.html @@ -101,6 +101,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -206,13 +209,13 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_compartment_metrics.html b/docs/reference/hemibrain_compartment_metrics.html index 3f2f4b4f..d149b9c6 100644 --- a/docs/reference/hemibrain_compartment_metrics.html +++ b/docs/reference/hemibrain_compartment_metrics.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -215,19 +218,19 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_cut.html b/docs/reference/hemibrain_cut.html index a66da1a0..d12f719c 100644 --- a/docs/reference/hemibrain_cut.html +++ b/docs/reference/hemibrain_cut.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_download_neurons.html b/docs/reference/hemibrain_download_neurons.html index 3bf362a8..474aa8ca 100644 --- a/docs/reference/hemibrain_download_neurons.html +++ b/docs/reference/hemibrain_download_neurons.html @@ -108,6 +108,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_extract_connections.html b/docs/reference/hemibrain_extract_connections.html index 903eebdf..a53866fd 100644 --- a/docs/reference/hemibrain_extract_connections.html +++ b/docs/reference/hemibrain_extract_connections.html @@ -101,6 +101,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_flow_centrality.html b/docs/reference/hemibrain_flow_centrality.html index baaeec2e..e534dc93 100644 --- a/docs/reference/hemibrain_flow_centrality.html +++ b/docs/reference/hemibrain_flow_centrality.html @@ -100,6 +100,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -239,10 +242,10 @@
- + hemibrain and flywire meta data +
- flywire @@ -192,7 +195,7 @@
"Hartenstein_Hemilineage" - the hemilineage that we reckon this cell type belongs to, based on expert review of light level data from the V. Hartenstein Lee group (Wong 2013, Lovick 2013). See
hemibrain_hemilineages."FAFB.match" - the ID of the manual match from the FAFB data set. ID indicates a neuron reconstructed in FAFBv14 CATMAID. Many of these neurons will be available through Virtual Fly Brain.
-"FAFB.match.quality" - the matcher makers qualitative assement of how good this match is: a poor match could be a neuron from a very similar same cell type or a highly untraced neuron that may be the correct cell type. An okay match should be a neuron that looks to be from the same morphological cell type but there may be some discrepancies in its arbour. A good match is a +
"FAFB.match.quality" - the matcher makers qualitative assessment of how good this match is: a poor match could be a neuron from a very similar same cell type or a highly untraced neuron that may be the correct cell type. An okay match should be a neuron that looks to be from the same morphological cell type but there may be some discrepancies in its arbour. A good match is a neuron that corresponds well between FAFB and the hemibrain data.
"layer" - probabilistic mean path length from neuron from ALRNs, depends on connection strengths.
"layer.ct"- the mean layer for cell type, rounded to the nearest whole number.
diff --git a/docs/reference/hemibrain_matched.html b/docs/reference/hemibrain_matched.html
index b4e06516..d1ee09c6 100644
--- a/docs/reference/hemibrain_matched.html
+++ b/docs/reference/hemibrain_matched.html
@@ -107,6 +107,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -183,14 +186,14 @@
"cell.type" - the neuPrint designated 'type' for the neuron. If
datasetis not"hemibrain", then this is based on the hemibrainmatch."cell" - the unique cell, which is just
cell.type#number."cellBodyFiber" - the cell body fiber to which this neuron belongs
"ItoLee_Hemilineage" - the hemilineage to which this neuron belongs. Seer
hemibrain_hemilineages."match" - the ID of the manual match from the other data set. If
dataset=="hemibrain"then this is aflywire.idthat can be found inflywire_neurons.If"CATMAID"or"flywire"then it is a hemibrain body ID.
-"quality" - the matcher makers qualitative assement of how good this match is.
+"quality" - the matcher makers qualitative assessment of how good this match is.
"FAFB.hemisphere.match" - the flywire coordinates of a neuron on the opposite hemisphere, which a match maker has designated as this
id's cognate."FAFB.hemisphere.match.quality" - the quality of this match.
"LM.match" - indicates a light level neuron that is a match for
id. This neuron will be inflycircuit_neurons()or other light level data.
diff --git a/docs/reference/hemibrain_matches.html b/docs/reference/hemibrain_matches.html
index 7ad49afe..c03832af 100644
--- a/docs/reference/hemibrain_matches.html
+++ b/docs/reference/hemibrain_matches.html
@@ -102,6 +102,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -193,14 +196,14 @@
"cell.type" - the neuPrint designated 'type' for the neuron. If
datasetis not"hemibrain", then this is based on the hemibrainmatch."cell" - the unique cell, which is just
cell.type#number."cellBodyFiber" - the cell body fiber to which this neuron belongs
"ItoLee_Hemilineage" - the hemilineage to which this neuron belongs. Seer
hemibrain_hemilineages."match" - the ID of the manual match from the other data set. If
dataset=="hemibrain"then this is aflywire.idthat can be found inflywire_neurons.If"CATMAID"or"flywire"then it is a hemibrain body ID.
-"quality" - the matcher makers qualitative assement of how good this match is.
+"quality" - the matcher makers qualitative assessment of how good this match is.
"FAFB.hemisphere.match" - the flywire coordinates of a neuron on the opposite hemisphere, which a match maker has designated as this
id's cognate."FAFB.hemisphere.match.quality" - the quality of this match.
"LM.match" - indicates a light level neuron that is a match for
id. This neuron will be inflycircuit_neurons()or other light level data.
diff --git a/docs/reference/hemibrain_matching.html b/docs/reference/hemibrain_matching.html
index c5111033..c78e1885 100644
--- a/docs/reference/hemibrain_matching.html
+++ b/docs/reference/hemibrain_matching.html
@@ -106,6 +106,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -214,7 +217,7 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_nblast.html b/docs/reference/hemibrain_nblast.html index 3450b95f..8ebc43a1 100644 --- a/docs/reference/hemibrain_nblast.html +++ b/docs/reference/hemibrain_nblast.html @@ -103,6 +103,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_olfactory_layers.html b/docs/reference/hemibrain_olfactory_layers.html index 83fa8b1d..cef28f32 100644 --- a/docs/reference/hemibrain_olfactory_layers.html +++ b/docs/reference/hemibrain_olfactory_layers.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_overlap_locality.html b/docs/reference/hemibrain_overlap_locality.html index dbdfd550..4347477c 100644 --- a/docs/reference/hemibrain_overlap_locality.html +++ b/docs/reference/hemibrain_overlap_locality.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_precomputed_splitpoints.html b/docs/reference/hemibrain_precomputed_splitpoints.html index 826008eb..76c52414 100644 --- a/docs/reference/hemibrain_precomputed_splitpoints.html +++ b/docs/reference/hemibrain_precomputed_splitpoints.html @@ -102,6 +102,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_read_neurons.html b/docs/reference/hemibrain_read_neurons.html index 2a5ea880..e49015e7 100644 --- a/docs/reference/hemibrain_read_neurons.html +++ b/docs/reference/hemibrain_read_neurons.html @@ -100,6 +100,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_remove_bad_synapses.html b/docs/reference/hemibrain_remove_bad_synapses.html index 4c076729..bc462b1b 100644 --- a/docs/reference/hemibrain_remove_bad_synapses.html +++ b/docs/reference/hemibrain_remove_bad_synapses.html @@ -102,6 +102,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -229,7 +232,7 @@
- + hemibrain and flywire meta data +
- flywire @@ -216,10 +219,10 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_skeleton_check.html b/docs/reference/hemibrain_skeleton_check.html index b5a91923..e430dcb1 100644 --- a/docs/reference/hemibrain_skeleton_check.html +++ b/docs/reference/hemibrain_skeleton_check.html @@ -100,6 +100,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -229,7 +232,7 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_splitpoints.html b/docs/reference/hemibrain_splitpoints.html index 584250a2..0de0aa38 100644 --- a/docs/reference/hemibrain_splitpoints.html +++ b/docs/reference/hemibrain_splitpoints.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_tags.html b/docs/reference/hemibrain_tags.html index 89824bc4..0abdd47b 100644 --- a/docs/reference/hemibrain_tags.html +++ b/docs/reference/hemibrain_tags.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -205,7 +208,7 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrain_use_splitpoints.html b/docs/reference/hemibrain_use_splitpoints.html index 36470401..ee604b92 100644 --- a/docs/reference/hemibrain_use_splitpoints.html +++ b/docs/reference/hemibrain_use_splitpoints.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -206,23 +209,23 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrainr-package.html b/docs/reference/hemibrainr-package.html index 7c4fd581..fe0c8f12 100644 --- a/docs/reference/hemibrainr-package.html +++ b/docs/reference/hemibrainr-package.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrainr_googledrive_data.html b/docs/reference/hemibrainr_googledrive_data.html index 33aa30ac..9c65cfe6 100644 --- a/docs/reference/hemibrainr_googledrive_data.html +++ b/docs/reference/hemibrainr_googledrive_data.html @@ -102,6 +102,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemibrainr_set_drive.html b/docs/reference/hemibrainr_set_drive.html index 9fb311dc..2752b8a9 100644 --- a/docs/reference/hemibrainr_set_drive.html +++ b/docs/reference/hemibrainr_set_drive.html @@ -101,6 +101,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/hemilineages.html b/docs/reference/hemilineages.html index 3dddd6df..050f3e24 100644 --- a/docs/reference/hemilineages.html +++ b/docs/reference/hemilineages.html @@ -97,6 +97,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/index.html b/docs/reference/index.html index c3aad38d..4006ffdf 100644 --- a/docs/reference/index.html +++ b/docs/reference/index.html @@ -95,6 +95,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -423,7 +426,7 @@
- + hemibrain and flywire meta data +
- flywire @@ -178,10 +181,10 @@
"Hartenstein_Hemilineage" - the hemilineage that we reckon this cell type belongs to, based on expert review of light level data from the V. Hartenstein Lee group (Wong 2013, Lovick 2013). See
hemibrain_hemilineages."FAFB.match" - the ID of the manual match from the FAFB data set. ID indicates a neuron reconstructed in FAFBv14 CATMAID. Many of these neurons will be available through Virtual Fly Brain.
-"FAFB.match.quality" - the matcher makers qualitative assement of how good this match is: a poor match could be a neuron from a very similar same cell type or a highly untraced neuron that may be the correct cell type. An okay match should be a neuron that looks to be from the same morphological cell type but there may be some discrepancies in its arbour. A good match is a +
"FAFB.match.quality" - the matcher makers qualitative assessment of how good this match is: a poor match could be a neuron from a very similar same cell type or a highly untraced neuron that may be the correct cell type. An okay match should be a neuron that looks to be from the same morphological cell type but there may be some discrepancies in its arbour. A good match is a neuron that corresponds well between FAFB and the hemibrain data.
"layer" - probabilistic mean path length from neuron from ALRNs, depends on connection strengths.
"layer.ct"- the mean layer for cell type, rounded to the nearest whole number.
@@ -220,7 +223,7 @@ "segregation_index"- a quantification of how polarised a neuron is, in terms of its segregation of inputs onto its predicted dendrite and outputs onto its axon, where 0 is no-polarisation and 1 is totally polarised (Schneider-Mizell 2016).
"notes"- other notes from annotators.
-"excpected_cit"a citation that describes this glomerulus,
+"expected_cit"a citation that describes this glomerulus,
"expected_RN_female_1h".
"expected_RN_female_SD".
"missing".
diff --git a/docs/reference/manually_assign_labels.html b/docs/reference/manually_assign_labels.html
index 737ca6e5..1f8aa8e9 100644
--- a/docs/reference/manually_assign_labels.html
+++ b/docs/reference/manually_assign_labels.html
@@ -98,6 +98,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -212,9 +215,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -148,7 +151,7 @@
Update the flywire.id column in a set of google sheets based on flywire xyz
-This function retreives flywire IDs based on xyz positions in flywire voxel space, from a set of google sheets. +
This function retrieves flywire IDs based on xyz positions in flywire voxel space, from a set of google sheets. It also writes the updated flywire IDs to the same google sheets. This is often helpful because flywire IDs are inherently unstable, they change every time a neuron is modified even slightly. Users can record 'stable' points in a neuron that identify it, e.g. a single xyz position in the cell body fibre, or at the soma, and then use this function to update and get the correct flywire ID whenever they wish.
@@ -160,6 +163,8 @@Update the flywire.id column in a set of google sheets based on flywire xyz chosen.columns = c("fw.x", "fw.y", "fw.z", "flywire.xyz", "flywire.id", "skid", "FAFB.xyz", "cell.type", "side", "ItoLee_Hemilineage", "Hartenstein_Hemilineage", "hemibrain_match"), + work_sheets = NULL, + meta = NULL, numCores = 1, max.tries = 10 ) @@ -174,8 +179,17 @@
Arg
chosen.columns -as well as writing column updates to the specified google sheets, this function returns a
data.framebuilt fromm all given sheets and their +as well as writing column updates to the specified google sheets, this function returns a
data.framebuilt from all given sheets and their individual tabs, that have been updated. This argument specifies which column you want returned. Filled with NAs if it does not exist.+ +work_sheets +a character vector of work sheet, i.e. tab, names for the googlesheet. If given, this function will only update those tabs.
+ meta +meta data for flywire neurons, e.g. as retreived using
flywire_meta. Used to efficiently inputflywire.xyzcolumn if only aflywire.identry has been given. +Only works if that id is also in this provideddata.frame,meta.numCores diff --git a/docs/reference/flywire_neurons.html b/docs/reference/flywire_neurons.html index 2ac758fe..f7492f7f 100644 --- a/docs/reference/flywire_neurons.html +++ b/docs/reference/flywire_neurons.html @@ -52,7 +52,8 @@ without loading the entire, large neuronlist into memory. You will need access to the hemibrain Google Team Drive and have it mounted with Google filestream.The function flywire_neurons_update can be used to update the available data. If you want to flag flywire neurons that should be added to the Google drive, without doing this yourself, you can use - flywire_request." /> + flywire_request. The flywire_basics function will calculate some useful meta-data, namely +a point in the primary neurite tract that can be used as a stable reference for this neuron." /> @@ -101,6 +102,9 @@-
+
Download a large set of well-traced skeletonised neurons from FlyWire
without loading the entire, largeneuronlistinto memory. You will need access to the hemibrain Google Team Drive and have it mounted with Google filestream.The functionflywire_neurons_updatecan be used to update the available data. If you want to flag flywire neurons that should be added to the Google drive, without doing this yourself, you can use -flywire_request. +flywire_request. Theflywire_basicsfunction will calculate some useful meta-data, namely +a point in the primary neurite tract that can be used as a stable reference for this neuron.flywire_neurons( @@ -181,6 +186,8 @@
Download a large set of well-traced skeletonised neurons from FlyWire
... ) +flywire_basics(flywire.neurons, ...) + flywire_request( request, selected_file = options()$flywire_flagged_gsheet, @@ -215,6 +222,11 @@Arg
nblast which flywire NBLAST to update on Google drive.
+ flywire.neurons +a
neuronlistof flywire neurons in FlyWire space. Theflywire_basicsfunction will calculate some useful meta-data, namely +a point in the primary neurite tract that can be used as a stable reference for this neuron.request a neuronlist, matrix of x,y,z position or flywire ID to add to a @@ -255,7 +267,31 @@
See a
Examples
diff --git a/docs/reference/glomerulus.html b/docs/reference/glomerulus.html index 2dff0754..0d1bc7b4 100644 --- a/docs/reference/glomerulus.html +++ b/docs/reference/glomerulus.html @@ -96,6 +96,9 @@# \donttest{ if (FALSE) { +# Loads all processed flywire neurons as a neuronlistfh object fw.neurons = flywire_neurons() + +# Get them already bridged to the JRC2018F brainsapce +## (Bogovic et al. 2018, high performance brain space) +fw.neurons.jrc2018f = flywire_neurons(brain = "JRC2018F") + +# Get them already mirrored to the other hemispehre, i.e. flipped +fw.neurons.jrc2018f.m = flywire_neurons(brain = "JRC2018F", mirror = TRUE) + +# Now say you have some flywire ID~s you wants to add +## to the nightly processing so they are available from Google drive. +## And they are stored in a .csv: +library(readr) +csv = read_csv("/Users/abates/Downloads/FlyWire_list.txt", +col_type = cols(.default = "c")) +ids = csv[,1][[1]] +neurons = skeletor(ids) +neurons.with.info = hemibrainr:::flywire_basics(neurons) +new.points = nat::xyzmatrix(neurons.with.info[,"flywire.xyz"]) +flywire_request(new.points) +## Now they are added to a google sheet, and will be read and +## processed as part of this nightly pipeline: +### https://github.com/flyconnectome/fafbpipeline + }# }-
+
-
+
-
+
-
+
Manage hemibrain-FAFB neuron matches
flywire_matching_rewrite( flywire.ids = names(flywire_neurons()), meta = flywire_neurons()[, ], + catmaid.update = TRUE, selected_file = options()$hemibrainr_matching_gsheet, + reorder = FALSE, + top.nblast = FALSE, + nblast = NULL, ... ) @@ -194,6 +201,8 @@
Manage hemibrain-FAFB neuron matches
fafb_matching_rewrite( selected_file = options()$hemibrainr_matching_gsheet, top.nblast = FALSE, + reorder = FALSE, + nblast = NULL, ... ) @@ -201,6 +210,9 @@Manage hemibrain-FAFB neuron matches
ids = NULL, selected_file = options()$hemibrainr_matching_gsheet, top.nblast = FALSE, + meta = NULL, + nblast.hemibrain.catmaid = NULL, + nblast.hemibrain.flywire = NULL, ... ) @@ -221,10 +233,26 @@Arg
meta meta data for the given flycircuit IDs.
+ catmaid.update +logical. Whether or not to update
flywire.xyzandflywire.idcolumns, based on e CATMAID neuron specified by askidcolumn.selected_file Specifies which Google Sheet to use. Unless you are using a personal Google Sheet, this should be
options()$hemibrainr_matching_gsheet.+ +reorder +logical. Whether or not to re-write the sheet so that it is ordered by hemilineage.
+ +top.nblast +logical. Whether or not to also give the top NBLAST match for each entry.
+ nblast +if
top.nblastisTRUEthis nblast matrix is used to update the columntop.nblast. If set toNULLdefaults to usinghemibrain_nblast. Columns should be hemibrain neurons, and rows the other data set.... arguments passed to methods for, for example,
neuprintr::neuprint_get_metaandelmr::fafb_get_meta.Arg
the tab to which to add your new information. You are either adding to information related to hemibrain neurons, or FAFB neurons.
- +top.nblast -logical. Whether or not to also give the top NBLAST match for each entry.
nblast.hemibrain.catmaid +if
top.nblastisTRUEthis nblast matrix is used to update the columntop.nblast. If set toNULLdefaults to usinghemibrain_nblast. Columns should be hemibrain neurons, and rows CATMAID neurons.+ nblast.hemibrain.flywire +if
top.nblastisTRUEthis nblast matrix is used to update the columntop.nblast. If set toNULLdefaults to usinghemibrain_nblast. Columns should be hemibrain neurons, and rows flywire neurons.flycircuit.ids diff --git a/docs/reference/hemibrain_adjust_saved_somas.html b/docs/reference/hemibrain_adjust_saved_somas.html index fd63a07a..4fb133fb 100644 --- a/docs/reference/hemibrain_adjust_saved_somas.html +++ b/docs/reference/hemibrain_adjust_saved_somas.html @@ -105,6 +105,9 @@-
+
-
+
-
+
Value
hemibrain_glomeruli_summaryis adata.framewith the columns:-
-
Details
note that hemibrain coordinate system has the anterior-posterior axis aligned with the Y axis (rather than the Z axis, which is more commonly observed).
-These meshes were generated by Sri Jagannathan and Tomke Stuermer based on +
These meshes were generated by Sri Jagannathan and Tomke Stuerner based on the location of PN dendritic synapses.
See also
diff --git a/docs/reference/hemibrain_cbf.html b/docs/reference/hemibrain_cbf.html index 7858af48..77235433 100644 --- a/docs/reference/hemibrain_cbf.html +++ b/docs/reference/hemibrain_cbf.html @@ -96,6 +96,9 @@-
+
-
+
Examp al.local.neurons=al.local.neurons[1:3] # Get neurons neurons = neuprintr::neuprint_read_neurons(al.local.neurons) - +
#> Error in neuprintr::neuprint_read_neurons(al.local.neurons): Error reading bodyids#> Warning: 2 neurons cropped, split likely to be inaccurate for: 2068966051, 2069311379#> Warning: 3 neurons have no soma tagged, split could be inaccurate for: 1702323386, 2068966051, 2069311379+#> Error in hemibrain_flow_centrality(neurons): object 'neurons' not found# Clean neurons neurons.cleaned = hemibrain_clean_skeleton(neurons) - +#> Error in hemibrain_clean_skeleton(neurons): object 'neurons' not foundif (FALSE) { # Plot the split to check it nat::nopen3d() diff --git a/docs/reference/hemibrain_colours.html b/docs/reference/hemibrain_colours.html index cdc4a50e..bd7878c6 100644 --- a/docs/reference/hemibrain_colours.html +++ b/docs/reference/hemibrain_colours.html @@ -107,6 +107,9 @@-
+
-
+
Examp # Get neurons neurons = neuprint_read_neurons(al.local.neurons) - +
#> Error in neuprint_read_neurons(al.local.neurons): Error reading bodyids#> Error in hemibrainr::scale_neurons(neurons): object 'neurons' not found#> Warning: 2 neurons cropped, split likely to be inaccurate for: 2068966051, 2069311379#> Warning: 3 neurons have no soma tagged, split could be inaccurate for: 1702323386, 2068966051, 2069311379+#> Error in hemibrain_flow_centrality(neurons): object 'neurons' not found# Calculate overlap and other metrics hcm = hemibrain_compartment_metrics(neurons, resample = NULL, just.leaves = TRUE, # just look how much the leaves of skeletons overlap, quicker (default) delta = 5) - +#> Error in is.neuronlist(X): object 'neurons' not found# }diff --git a/docs/reference/hemibrain_connectivity_similarity.html b/docs/reference/hemibrain_connectivity_similarity.html index ce28701f..96244b49 100644 --- a/docs/reference/hemibrain_connectivity_similarity.html +++ b/docs/reference/hemibrain_connectivity_similarity.html @@ -96,6 +96,9 @@-
+
-
+
-
+
-
+
-
+
Examp # Get neurons neurons = neuprintr::neuprint_read_neurons(exemplars) - +
#> Error in neuprintr::neuprint_read_neurons(exemplars): Error reading bodyids# Now use a pre-saved axon-dendrite split neurons.flow = hemibrain_flow_centrality(neurons) - +#> Error in hemibrain_flow_centrality(neurons): object 'neurons' not foundif (FALSE) { # Plot the split to check it nat::nopen3d() diff --git a/docs/reference/hemibrain_get_meta.html b/docs/reference/hemibrain_get_meta.html index 66e8c372..cb6fcc40 100644 --- a/docs/reference/hemibrain_get_meta.html +++ b/docs/reference/hemibrain_get_meta.html @@ -98,6 +98,9 @@-
+
Value
-
+
FormatValue
a
data.framewhere each row is a neuron, either from the hemibrain or FAFB data sets. Each row gives you its matching neuron in the other data set. These matches have been -manually assigned usingfafb_matching,hemibrain_matchingandLR_matching. If you use this information make sure you credit it appriopriately. +manually assigned usingfafb_matching,hemibrain_matchingandLR_matching. If you use this information make sure you credit it appropriately. Contact us if unsure:-
+
Value
a
data.framewhich includes neuron's ID (either its CATMAID skeleton ID or neuprint body ID), the data set from which it comes, its putative cell type and connectivity type, and its match in the other dataset.a
data.framewhere each row is a neuron, either from the hemibrain or FAFB data sets. Each row gives you its matching neuron in the other data set. These matches have been -manually assigned usingfafb_matching,hemibrain_matchingandLR_matching. If you use this information make sure you credit it appriopriately. +manually assigned usingfafb_matching,hemibrain_matchingandLR_matching. If you use this information make sure you credit it appropriately. Contact us if unsure:-
+
Match up neurons between the hemibrain and FAFB
hemibrain.nblast = NULL, threshold = 0, selected_file = options()$hemibrainr_matching_gsheet, - batch_size = 50, + batch_size = 20, db = hemibrain_neurons(brain = "FAFB14"), query = NULL, overwrite = c("FALSE", "mine", "mine_empty", "TRUE"), diff --git a/docs/reference/hemibrain_metrics.html b/docs/reference/hemibrain_metrics.html index 649f33f6..1be8e360 100644 --- a/docs/reference/hemibrain_metrics.html +++ b/docs/reference/hemibrain_metrics.html @@ -103,6 +103,9 @@-
+
-
+
-
+
-
+
-
+
-
+
-
+
Examp
# \donttest{ # Read in a problematic neuron neuron = neuprint_read_neurons(5813020793) - +#> Error in neuprint_read_neurons(5813020793): Error reading bodyidsif (FALSE) { library(nat) plot3d(neuron) @@ -238,7 +241,7 @@Examp # remove any synapses outside of this surface (or on the cell body fibre) neuron.fixed = hemibrain_remove_bad_synapses(neuron, meshes=hemibrain.surf) -if (FALSE) { +
#> Error in UseMethod("hemibrain_remove_bad_synapses"): no applicable method for 'hemibrain_remove_bad_synapses' applied to an object of class "function"if (FALSE) { # previously we used all the individual hemibrain meshes. This may produce # slightly different results but using *hemibrain.surf* is much faster neuron.fixed = hemibrain_remove_bad_synapses(neuron, meshes=hemibrain_roi_meshes()) diff --git a/docs/reference/hemibrain_reroot.html b/docs/reference/hemibrain_reroot.html index 5149deb3..9da09092 100644 --- a/docs/reference/hemibrain_reroot.html +++ b/docs/reference/hemibrain_reroot.html @@ -102,6 +102,9 @@-
+
Examp # Read in these neurons neurons.bs = neuprintr::neuprint_read_neurons(bad.soma) - +
#> Error in neuprintr::neuprint_read_neurons(bad.soma): Error reading bodyids# Re-root neurons = hemibrain_reroot(neurons.bs, meshes = hemibrain.surf) - +#> Error in hemibrain_reroot(neurons.bs, meshes = hemibrain.surf): object 'neurons.bs' not foundif (FALSE) { # Let's check that this worked. Old root in red, new in green nat::nopen3d() diff --git a/docs/reference/hemibrain_roi_meshes.html b/docs/reference/hemibrain_roi_meshes.html index 888f53f0..e750288e 100644 --- a/docs/reference/hemibrain_roi_meshes.html +++ b/docs/reference/hemibrain_roi_meshes.html @@ -100,6 +100,9 @@-
+
-
+
Examp # Read in these neurons neurons = neuprintr::neuprint_read_neurons(ids) -if (FALSE) { +
#> Error in neuprintr::neuprint_read_neurons(ids): Error reading bodyidsif (FALSE) { # Get all the roi meshes hemibrain.rois = hemibrain_roi_meshes() ## Using this as the argument for 'meshes' will also @@ -243,7 +246,7 @@Examp # whole hemibrain neurons.checked = hemibrain_skeleton_check(neurons, meshes = hemibrain.surf) -
#> Re-rooting 1 neurons without a soma#> Removing synapses outside the hemibrain neuropil volume for 3 neurons#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated+#> Error in nat::as.neuronlist(x): object 'neurons' not foundif (FALSE) { # Let's check that this worked nat::nopen3d() diff --git a/docs/reference/hemibrain_somas.html b/docs/reference/hemibrain_somas.html index b1185a14..4f40500a 100644 --- a/docs/reference/hemibrain_somas.html +++ b/docs/reference/hemibrain_somas.html @@ -101,6 +101,9 @@-
+
-
+
-
+
Examp # Get neurons neurons = hemibrain_read_neurons(al.local.neurons) -
#> Using Google Team Drive: hemibrainr#> Warning: 1 neurons cropped, split likely to be inaccurate for: 2068966051#> Warning: 1 neurons have no soma tagged, split could be inaccurate for: 2068966051#> Error in `$<-.data.frame`(`*tmp*`, "class", value = NA): replacement has 1 row, data has 0+#> Using Google Team Drive: hemibrainr#> Error in neuprintr::neuprint_read_neurons(x, ...): Error reading bodyids# Tag neurons = hemibrain_settags(neurons, antennal.lobe = TRUE)#> Error in hemibrain_settags(neurons, antennal.lobe = TRUE): object 'neurons' not founddiff --git a/docs/reference/hemibrain_type_plot.html b/docs/reference/hemibrain_type_plot.html index 6524f880..f0df731e 100644 --- a/docs/reference/hemibrain_type_plot.html +++ b/docs/reference/hemibrain_type_plot.html @@ -97,6 +97,9 @@-
+
-
+
Examp # Get neurons neurons = neuprintr::neuprint_read_neurons(tough) - +
#> Error in neuprintr::neuprint_read_neurons(tough): Error reading bodyids# Now make sure the neurons have a soma marked ## Some hemibrain neurons do not, as the soma was chopped off neurons.checked = hemibrain_skeleton_check(neurons, meshes = hemibrain.surf) -#> Re-rooting 2 neurons without a soma#> Removing synapses outside the hemibrain neuropil volume for 5 neurons#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated#> Warning: invalid factor level, NA generated+#> Error in nat::as.neuronlist(x): object 'neurons' not found# Split neuron ## These are the recommended parameters for hemibrain neurons neurons.flow = flow_centrality(neurons.checked, polypre = TRUE, mode = "centrifugal", split = "distance") - +#> Error in flow_centrality(neurons.checked, polypre = TRUE, mode = "centrifugal", split = "distance"): object 'neurons.checked' not found#> Error in nat::is.neuronlist(x): object 'neurons.flow' not found# Re-use the results neurons.flow.2 = hemibrain_use_splitpoints(neurons, splitpoints, knn = FALSE) -#> Error in if (is.na(dendrite.primary)) { dendrite.primary = dendrite.start[1]}: argument is of length zero+#> Error in hemibrain_use_splitpoints(neurons, splitpoints, knn = FALSE): object 'neurons' not foundif (FALSE) { # Plot the split to check it nat::nopen3d() diff --git a/docs/reference/hemibrain_view.html b/docs/reference/hemibrain_view.html index f5a5414c..be36c286 100644 --- a/docs/reference/hemibrain_view.html +++ b/docs/reference/hemibrain_view.html @@ -96,6 +96,9 @@-
+
-
+
-
+
-
+
-
+
-
+
-
+rn.idsorn.idshrn.idspn.idsupn.idsmpn.idsvppn.idsdan.idsmbon.idsalln.idston.idslhn.idsdn.idskc.idsapl.idscent.idslc.idskc.idsrn.inforn.idsorn.idshrn.idspn.idsupn.idsmpn.idsvppn.idsdan.idsmbon.idsalln.idston.idslhn.idsdn.idskc.idsapl.idscent.idslc.idsrn.infoBodyids for neuron classes
@@ -435,7 +438,7 @@ -
+standard_transmitters()standard_lineages()standard_compartments()standardise()standard_transmitters()standard_statuses()standard_lineages()standard_compartments()standardise()Use standard names and spellings
@@ -652,13 +655,13 @@ flywire_neurons()
flywire_neurons_update()flywire_nblast_update()flywire_request()+flywire_neurons()flywire_neurons_update()flywire_nblast_update()flywire_basics()flywire_request()Download a large set of well-traced skeletonised neurons from FlyWire
-
+flywire_meta()flywire_contributions()flywire_ids()flywire_meta()flywire_failed()flywire_contributions()flywire_ids()Read precomputed flywire data from the hemibrainr Google Drive
diff --git a/docs/reference/info.html b/docs/reference/info.html index b0542645..3d122abd 100644 --- a/docs/reference/info.html +++ b/docs/reference/info.html @@ -99,6 +99,9 @@ -
+
FormatAn object of class
data.framewith 70 rows and 58 columns.An object of class
data.framewith 2383 rows and 48 columns.An object of class
-data.framewith 2129 rows and 45 columns.An object of class
+data.framewith 197 rows and 47 columns.An object of class
data.framewith 197 rows and 48 columns.An object of class
data.framewith 1927 rows and 45 columns.An object of class
-data.framewith 24 rows and 45 columns.An object of class
+data.framewith 431 rows and 33 columns.An object of class
data.framewith 345 rows and 33 columns.Details
@@ -205,7 +208,7 @@Details
Details
-
+
Examp "326530038", "203253253", "5813079341") neurons = neuprintr::neuprint_read_neurons(exemplars) - +
#> Error in neuprintr::neuprint_read_neurons(exemplars): Error reading bodyids#> Error in hemibrain_flow_centrality(neurons): object 'neurons' not foundif (FALSE) { # Plot the split to check it, correcting any errors nat::nopen3d() diff --git a/docs/reference/matches_update.html b/docs/reference/matches_update.html index 197a33da..b2bbc096 100644 --- a/docs/reference/matches_update.html +++ b/docs/reference/matches_update.html @@ -47,7 +47,7 @@ - @@ -98,6 +98,9 @@-
+
Update neuron match information on a google sheet
-@@ -194,7 +197,7 @@This function retreives neuron matches (
hemibrain_matches) from a master-matching google sheet. If then +This function retrieves neuron matches (
hemibrain_matches) from a master-matching google sheet. If then can update other google sheets, specified by the user to update neuron-match information. Just columns giving the match, match quality and cell type are updated.Arg
chosen.columns -as well as writing column updates to the specified google sheets, this function returns a
data.framebuilt fromm all given sheets and their +as well as writing column updates to the specified google sheets, this function returns a
data.framebuilt from all given sheets and their individual tabs, that have been updated. This argument specifies which column you want returned. Filled with NAs if it does not exist.-
+
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/plot3d_split.html b/docs/reference/plot3d_split.html index a954e872..d8f720cf 100644 --- a/docs/reference/plot3d_split.html +++ b/docs/reference/plot3d_split.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/plot_inspirobot.html b/docs/reference/plot_inspirobot.html index f2a5cbfd..d4231f61 100644 --- a/docs/reference/plot_inspirobot.html +++ b/docs/reference/plot_inspirobot.html @@ -98,6 +98,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/primary_neurite.html b/docs/reference/primary_neurite.html index 98bf1e46..90902070 100644 --- a/docs/reference/primary_neurite.html +++ b/docs/reference/primary_neurite.html @@ -103,6 +103,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -235,10 +238,10 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/reexports.html b/docs/reference/reexports.html index e6188db5..8bedca62 100644 --- a/docs/reference/reexports.html +++ b/docs/reference/reexports.html @@ -105,6 +105,9 @@
- + hemibrain and flywire meta data +
- flywire diff --git a/docs/reference/standardise.html b/docs/reference/standardise.html index 21bf0886..9c3f90f0 100644 --- a/docs/reference/standardise.html +++ b/docs/reference/standardise.html @@ -96,6 +96,9 @@
- + hemibrain and flywire meta data +
- flywire @@ -151,6 +154,8 @@
-
+
-
+
-
+
Examp
#> Error in neuprintr::neuprint_read_neurons("451987038"): Error reading bodyids# Extract primary neurite pnt = primary_neurite(neuron) - +#> Error in UseMethod("primary_neurite"): no applicable method for 'primary_neurite' applied to an object of class "function"if (FALSE) { # Plot the primary neurite nat::nopen3d() diff --git a/docs/reference/prune_vertices.neuprintneuron.html b/docs/reference/prune_vertices.neuprintneuron.html index 41ed419b..989840f1 100644 --- a/docs/reference/prune_vertices.neuprintneuron.html +++ b/docs/reference/prune_vertices.neuprintneuron.html @@ -96,6 +96,9 @@-
+
-
+
-
+
Use standard names and spellings
standard_transmitters(x) +standard_statuses(x, invert = FALSE) + standard_lineages(x) standard_compartments(x, invert = FALSE) diff --git a/inst/images/hemibrainr_googledrive.png b/inst/images/hemibrainr_googledrive.png new file mode 100644 index 00000000..129152bb Binary files /dev/null and b/inst/images/hemibrainr_googledrive.png differ diff --git a/vignettes/google_filestream.Rmd b/vignettes/google_filestream.Rmd index 082b246c..0b1c118e 100644 --- a/vignettes/google_filestream.Rmd +++ b/vignettes/google_filestream.Rmd @@ -29,6 +29,10 @@ The best option is to use google filestream. By default, this is what `hemibrain We have two [Google team drives](https://support.google.com/a/users/answer/9310156?hl=en) available for you to use, which contain similar data. One (`"hemibrain"`) is for internal use by the [Drosophila Connectomics Group](https://www.zoo.cam.ac.uk/research/groups/connectomics). The other one (`"hemibrainr"`) is shared with those who would like access. Contact us by email to request access. +
+ + + ## Option 1: Google workspace, hemibrainr and R [Google drive](https://en.wikipedia.org/wiki/Google_Drive) is a data storage and synchronisation system. A Google team drive (shared drive) is a Google drive that is shared by a team of people, you can find them on your 'shared drives' in your Google authenticated account. Sharing and file ownership are managed for the entire team rather than one individual. Google Drive is a key component of [Google Workspace](https://workspace.google.com/pricing.html), Google's monthly subscription offering for businesses and organizations. As part of select Google Workspace plans, Drive offers unlimited storage, advanced file audit reporting, enhanced administration controls, and greater collaboration tools for teams.
