-
Notifications
You must be signed in to change notification settings - Fork 12
/
Copy path08_big_data.Rmd
598 lines (393 loc) · 16.7 KB
/
08_big_data.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
# Big data {#big_data}
```{r include = FALSE}
# Caching this markdown file
#knitr::opts_chunk$set(cache = TRUE)
```
## The Big Picture
- Big data problem: data is too big to fit into memory (=local environment).
- R reads data into random-access memory (RAM) at once, and this object lives in memory entirely. So, if object.size > memory.size, the process will crash R.
- Therefore, the key to dealing with big data in R is reducing the size of data you want to bring into it.
**Techniques to deal with big data**
- Medium-sized file (1-2 GB)
- Try to reduce the size of the file using slicing and dicing
- Tools:
- R:`data.table::fread(file path, select = c("column 1", "column 2"))`. This command imports data faster than `read.csv()` does.
- Command-line: [`csvkit`](https://csvkit.readthedocs.io/en/latest/) - a suite of command-line tools to and working with CSV
- Large file (> 2-10 GB)
- Put the data into a database and **ACCESS** it
- Explore the data and pull the objects of interest
**Databases**
- Types of databases
- Relational database = a **collection** of **tables** (fixed columns and rows): SQL is a staple tool to define, **query** (the focus of the workshop today), control, and manipulate this type of database
- Non-relational database = a collection of documents (MongoDB), key-values (Redis and DyanoDB), wide-column stores (Cassandra and HBase), or graph (Neo4j and JanusGraph). Note that this type of database does not preclude SQL. NoSQL stands for ["not only SQL."](https://www.mongodb.com/nosql-explained)
**Relational database example**

## SQL
- Structured Query Language. Called SEQUEL and was developed by IBM Corporation in the 1970s.
- Remains the standard language for a relational database management system.
- It's a DECLARATIVE language ([what to do > how to do](https://www.sqlite.org/queryplanner.html))
- Database management systems figure an optimal way to execute a query (query optimization)
```sql
SELECT COLUMN FROM TABLE
```
### Learning objectives
* Embracing a new mindset: shifting from ownership (opening CSVs stored in your laptop) to access (accessing data stored in a database)
* Learning how to use R and SQL to access and query a database
### SQL and R
* SQL and R
SQL | R
------------- | --------------------------------------------------------------------------
SELECT | select() for columns, mutate() for expressions, summarise() for aggregates
FROM | which data frame
WHERE | filter()
GROUP BY | group_by()
HAVING | filter() **after group_by()**
ORDER BY | arrange()
LIMIT | head()
**Challenge 1**
1. Can you tell me the difference in the order in which the following `R` and `SQL` codes were written to manipulate data? For instance, in R, what command comes first? In contrast, in SQL, what command comes first?
- R example
```r
data %>% # Data
select() %>% # Column
filter() %>% # Row
group_by() %>% # Group by
summarise(n = n()) %>% # n() is one of the aggregate functions in r; it's count() used inside summarise() function
filter() %>% # Row
order_by() # Arrange
```
- SQL example (in a SQL chunk, use `--` instead of `#` to comment)
```sql
SELECT column, aggregation (count())` -- Column
FROM data # Data
WHERE condition -- Filter rows
GROUP BY column -- Group by
HAVING condition -- Filter rows after group by
ORDER BY column -- Arrange
```
](https://wizardzines.com/zines/sql/samples/from.png)
### Setup
Let's get to work.
### Packages
- `pacman::p_load()` reduces steps for installing and loading several packages simultaneously.
```{r}
# pacman
if (!require("pacman")) install.packages("pacman")
# The rest of pkgs
pacman::p_load(
tidyverse, # tidyverse packages
DBI, # using SQL queries
RSQLite, # SQLite
dbplyr, # use database with dplyr
glue, # glue to automate workflow
nycflights13 # toy data
)
```
### NYC flights data
- [The flight on-time performance data](https://www.transtats.bts.gov/DL_SelectFields.asp?Table_ID=236) from the Bureau of Transportation Statistics of the U.S. government. The data goes back to 1987, and its size is more than 20 gigabytes. For practice, we only use a small subset of the original data (flight data departing NYC in 2013) provided by RStudio.
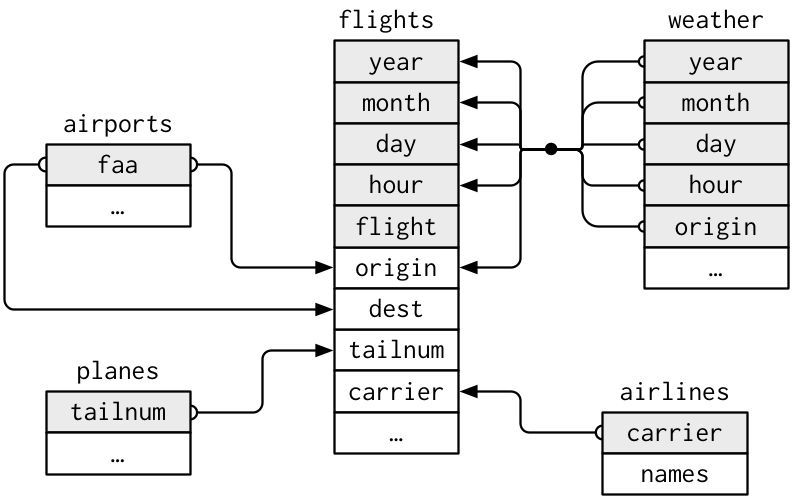
### Workflow
1. Create/connect to a database
- Note that the server also can be your laptop (called [localhost](https://en.wikipedia.org/wiki/Localhost#:~:text=In%20computer%20networking%2C%20localhost%20is,via%20the%20loopback%20network%20interface.)).
- Short answer: To do so, you need interfaces between R and a database. We use [`RSQLite`](https://github.com/r-dbi/RSQLite) in this tutorial because it's easy to set up.
- Long answer: The `DBI` package in R provides a client-side interface that allows `dplyr` to work with databases. DBI is automatically installed when you install `dbplyr`. However, you need to install a specific backend engine (a tool for communication between R and a database management system) for the database (e.g., `RMariaDB`, `RPostgres`, `RSQLite`). In this workshop, we use SQLite because it is the easiest to get started with. I love PostgreSQL because it's open-source and also powerful to do [many amazing things](https://www.postgresql.org/docs/current/functions.html) (e.g., text mining, geospatial analysis). If you want to build a data warehouse, an analytical platform, consider using Spark (Hadoop).
2. Copy a table to the database
- Option 1: You can create a table and insert rows manually. You also need to define the data schema (the database structure) to do that.
- Table
- Collection of rows
- Collection of columns (fields or attributes)
- Each col has a type:
- String: `VARCHAR(20)`
- Integer: `INTEGER`
- Floating-point: `FLOAT`, `DOUBLE`
- Date/time: `DATE`, `TIME`, `DATETIME`
- **Schema**: the structure of the database
- The table name
- The names and types of its columns
- Various optional additional information
- [Constraints](https://www.w3schools.com/sql/sql_constraints.asp)
- Syntax: `column datatype constraint`
- Examples: `NOT NULL`, `UNIQUE`, `INDEX`
```sql
-- Create table
CREATE TABLE students (
id INT AUTO_INCREMENT,
name VARCHAR(30),
birth DATE,
gpa FLOAT,
grad INT,
PRIMARY KEY(id));
-- Insert one additional row
INSERT INTO students(name, birth, gpa, grad)
VALUES ('Adam', '2000-08-04', 4.0, 2020);
```
- Option 2: Copy a file (object) to a table in a database using `copy_to`). We take this option as it's fast, and we would like to focus on querying in this workshop.
3. Query the table
- Main focus
4. Pull the results of interests (**data**) using `collect()`
5. Disconnect the database
#### Create a database
```{r}
# Define a backend engine
drv <- RSQLite::SQLite()
# Create an empty in-memory database
con <- DBI::dbConnect(drv,
dbname = ":memory:")
# Connect to an existing database
#con <- DBI::dbConnect(RMariaDB::MariaDB(),
# host = "database.rstudio.com",
# user = "hadley",
# password = rstudioapi::askForPassword("Database password")
#)
dbListTables(con)
# character(0) = NULL
```
- Note that `con` is empty at this stage.
#### Copy an object as a table to the database (push)
```{r}
# Copy objects to the data
# copy_to() comes from dplyr
copy_to(dest = con,
df = flights)
copy_to(dest = con,
df = airports)
copy_to(dest = con,
df = planes)
copy_to(dest = con,
df = weather)
# If you need, you can also select which columns you would like to copy:
# copy_to(dest = con,
# df = flights,
# name = "flights",
# indexes = list(c("year", "tailnum", "dest")))
```
```{r basic information on tables and fields}
# Show two tables in the database
dbListTables(con)
# Show the columns/attributes/fields of a table
dbListFields(con, "flights")
dbListFields(con, "weather")
```
#### Quick demonstrations:
- SELECT desired columns
- FROM tables
- Select all columns (*) from `flights` table and show the `first ten rows`
- Note that you can combine SQL and R commands thanks to `dbplyr.`
- Option 1
```{r}
DBI::dbGetQuery(con,
"SELECT * FROM flights;") %>% # SQL
head(10) # dplyr
```
- Option 2 (works faster)
```{sql connection=con, eval=FALSE}
SELECT *
FROM flights
LIMIT 10
```
- Option 3 (automating workflow)
- When local variables are updated, the SQL query is also automatically updated. This approach is called [parameterized query](https://www.php.net/manual/en/pdo.prepared-statements.php) (or prepared statement).
```{r}
######################## PREPARATION ########################
# Local variables
tbl <- "flights"
var <- "dep_delay"
num <- 10
# Glue SQL query string
# Note that to indicate a numeric value, you don't need.
sql_query <- glue_sql("
SELECT {`var`}
FROM {`tbl`}
LIMIT {num}
", .con = con)
######################## EXECUTION ########################
# Run the query
dbGetQuery(con, sql_query)
```
**Challenge 2**
Can you rewrite the above code using `LIMIT` instead of `head(10)`?
- You may notice that using only SQL code makes querying faster.
- Select `dep_delay` and `arr_delay` from flights table, show the first ten rows, then turn the result into a tibble.
**Challenge 3**
Could you remind me how to see the list of attributes of a table? Let's say you want to see the `flights` table attributes. How can you do it?
- Collect the selected columns and filtered rows
```{r}
df <- dbGetQuery(con,
"SELECT dep_delay, arr_delay FROM flights;") %>%
head(10) %>%
collect()
```
- Counting rows
- Count all (*)
```{r}
dbGetQuery(con,
"SELECT COUNT(*)
FROM flights;")
```
```{r}
dbGetQuery(con,
"SELECT COUNT(dep_delay)
FROM flights;")
```
- Count distinct values
```{r}
dbGetQuery(con,
"SELECT COUNT(DISTINCT dep_delay)
FROM flights;")
```
#### Tidy-way: dplyr -> SQL
Thanks to the `dbplyr` package, you can use the `dplyr` syntax to query SQL.
- Note that pipe (%) works.
```{r}
# tbl select tables
flights <- con %>% tbl("flights")
airports <- con %>% tbl("airports")
planes <- con %>% tbl("planes")
weather <- con %>% tbl("weather")
```
- `select` = `SELECT`
```{r}
flights %>%
select(contains("delay"))
```
**Challenge 4**
Your turn: write the same code in SQL. Don't forget to add the `connection` argument to your SQL code chunk.
- `mutate` = `SELECT` `AS`
```{r}
flights %>%
select(distance, air_time) %>%
mutate(speed = distance / (air_time / 60))
```
**Challenge 5**
Your turn: write the same code in SQL. (
Hint: `mutate(new_var = var 1 * var2` (R) = `SELECT var1 * var2 AS near_var` (SQL)
- `filter` = `WHERE`
```{r}
flights %>%
filter(month == 1, day == 1) # filter(month ==1 & day == 1) Both work in the same way.
```
**Challenge 6**
Your turn: write the same code in SQL (hint: `filter(condition1, condition2)` = `WHERE condition1 and condition2`)
**Additional tips**
Note that R and SQL operators are not exactly alike. R uses `!=` for `Not equal to`. SQL uses `<>` or `!=`. Furthermore, there are some cautions about using `NULL` (NA; unknown or missing): it should be `IS NULL` or `IS NOT NULL` not `=NULL` or `!=NULL` (this makes sense because NULL represents an absence of a value).
Another pro-tip is [`LIKE` operator](https://www.w3schools.com/sql/sql_like.asp), used in a `WHERE` statement to find values based on string patterns.
```{sql, connection = con}
SELECT DISTINCT(origin) -- Distinct values from origin column
FROM flights
WHERE origin LIKE 'J%'; -- Find any origin values that start with "J"
```
`%` is one of the wildcards you can use for string matching. `%` matches any number of characters. So, `J%` matches Jae, JFK, Joseph, etc. `_` is another useful wildcard that matches exactly one character. So `J_` matches only JA, JE, etc. If wildcards are not enough, then you should consider using regular expressions.
- `arrange` = `ORDER BY`
```{r}
flights %>%
arrange(carrier, desc(arr_delay)) %>%
show_query()
```
**Challenge 7**
Your turn: write the same code in SQL.
Hint: `arrange(var1, desc(var2)` (R) = `ORDER BY var1, var2 DESC` (SQL)
- `summarise` = `SELECT` `AS` and `group by` = `GROUP BY`
```{r}
flights %>%
group_by(month, day) %>%
summarise(delay = mean(dep_delay))
```
**Challenge 8**
Your turn: write the same code in SQL (hint: in SQL the order should be `SELECT group_var1, group_var2, AVG(old_var) AS new_var` -> `FROM` -> `GROUP BY`)
- If you feel too much challenged, here's a help.
```{r}
flights %>%
group_by(month, day) %>%
summarise(delay = mean(dep_delay)) %>%
show_query() # Show the SQL equivalent!
```
- Joins
- Using joins is more straightforward in R than it is in SQL.
- However, more flexible joins exist in SQL, and they are not available in R.
- Joins involving 3+ tables are not supported.
- Some advanced joins available in SQL are not supported.
- For more information, check out [`tidyquery`](https://github.com/ianmcook/tidyquery/issues) to see the latest developments.
- SQL command
`FROM one table LEFT JOIN another table ON condition = condition` (`ON` in SQL = `BY` in R)
```{sql, connection = con}
SELECT *
FROM flights AS f
LEFT JOIN weather AS w
ON f.year = w.year AND f.month = w.month
```
Can anyone explain why SQL query using `dplyr` then translated by `show_query()` looks more complex than the above? ([Hint](https://stackoverflow.com/questions/36808295/how-to-remove-duplicate-columns-from-join-in-sql))
```{r}
flights %>%
left_join(weather, by = c("year", "month")) %>%
show_query()
```
#### Collect (pull)
* `collect()` is used to pull the data. Depending on the data size, it may take a long time to run.
- The following code won't work.
> Error in UseMethod("collect") : no applicable method for 'collect' applied to an object of class "c('LayerInstance', 'Layer', 'ggproto', 'gg')"
```{r eval = FALSE}
origin_flights_plot <- flights %>%
group_by(origin) %>%
tally() %>%
ggplot() +
geom_col(aes(x = origin, y = n)) %>%
collect()
```
- This works.
```{r}
df <- flights %>%
group_by(origin) %>%
tally() %>%
collect()
origin_flights_plot <- ggplot(df) +
geom_col(aes(x = origin, y = n))
origin_flights_plot
```
#### Disconnect
```{r}
DBI::dbDisconnect(con)
```
### Things we didn't cover
#### Subquery
Subquery = a query nested inside a query
This hypothetical example is inspired by [dofactory blog post](https://www.dofactory.com/sql/subquery).
```{sql, eval = FALSE}
SELECT names -- Outer query
FROM consultants
WHERE Id IN (SELECT ConsultingId
FROM consulting_cases
WHERE category = 'r' AND category = 'sql'); -- Subquery
```
#### Common table expression (WITH clauses)
This is just a hypothetical example inspired by [James LeDoux's blog post](https://jamesrledoux.com/code/sql-cte-common-table-expressions.
```{sql, eval = FALSE}
-- cases about R and SQL from dlab-database
WITH r_sql_consulting_cases AS ( -- The name of the CTE expression
-- The CTE query
SELECT
id
FROM
dlab
WHERE
tags LIKE '%sql%'
AND
tags LIKE '%r%'
),
-- count the number of open cases about this consulting category
-- The outer query
SELECT status, COUNT(status) AS open_status_count
FROM dlab as d
INNER JOIN r_sql_consulting_cases as r
ON d.id = r.id
WHERE status = 'open';
```
### References
- [csv2db](https://github.com/csv2db/csv2db) - for loading large CSV files in to a database
- R Studio, [Database using R](https://db.rstudio.com/)
- Ian Cook, ["Bridging the Gap between SQL and R"](https://github.com/ianmcook/rstudioconf2020/blob/master/bridging_the_gap_between_sql_and_r.pdf) rstudio::conf 2020 slides
- [Video recording](https://www.youtube.com/watch?v=JwP5KdWSgqE&ab_channel=RStudio)
- Data Carpentry contributors, [SQL database and R](https://datacarpentry.org/R-ecology-lesson/05-r-and-databases.html), Data Carpentry, September 10, 2019.
- [Introduction to dbplyr](https://cran.r-project.org/web/packages/dbplyr/vignettes/dbplyr.html)
- Josh Erickson, [SQL in R](http://dept.stat.lsa.umich.edu/~jerrick/courses/stat701/notes/sql.html), STAT 701, University of Michigan
- [SQL zine](https://wizardzines.com/zines/sql/) by Julia Evans
- [q](http://harelba.github.io/q/) - a command-line tool that allows direct execution of SQL-like queries on CSVs/TSVs (and any other tabular text files)