diff --git a/docs/no_toc/01-Fundamentals.md b/docs/no_toc/01-Fundamentals.md
index fe8e77e..2611821 100644
--- a/docs/no_toc/01-Fundamentals.md
+++ b/docs/no_toc/01-Fundamentals.md
@@ -4,11 +4,11 @@
## Goals of this course
-- Continue building *programming fundamentals*: how to make use of complex data structures, use custom functions built by other R users, and creating your own functions. How to iterate repeated tasks that scales naturally.
+- Continue building *programming fundamentals*: How to use complex data structures, use and create custom functions, and how to iterate repeated tasks
- Continue exploration of *data science fundamentals*: how to clean messy data to a Tidy form for analysis.
-- Outcome: Conduct a full analysis in the data science workflow (minus model).
+- At the end of the course, you will be able to: conduct a full analysis in the data science workflow (minus model).
{width="450"}
@@ -428,7 +428,7 @@ l1$score
Therefore, `l1$score` is the same as `l1[[4]]` and is the same as `l1[["score"]]`.
-A dataframe is just a named list of vectors of same length with **attributes** of (column) `names` and `row.names`.
+A dataframe is just a named list of vectors of same length with additional **attributes** of (column) `names` and `row.names`.
## Matrix
@@ -475,3 +475,7 @@ my_matrix[2, 3]
```
## [1] 6
```
+
+## Exercises
+
+You can find [exercises and solutions on Posit Cloud](https://posit.cloud/content/8236252), or on [GitHub](https://github.com/fhdsl/Intermediate_R_Exercises).
diff --git a/docs/no_toc/02-Data_cleaning_1.md b/docs/no_toc/02-Data_cleaning_1.md
index f5c9180..a90d9a0 100644
--- a/docs/no_toc/02-Data_cleaning_1.md
+++ b/docs/no_toc/02-Data_cleaning_1.md
@@ -158,7 +158,7 @@ grade2 = if_else(grade > 60, TRUE, FALSE)
3. If-else_if-else
-```
+```
grade3 = case_when(grade >= 90 ~ "A",
grade >= 80 ~ "B",
grade >= 70 ~ "C",
@@ -199,7 +199,7 @@ simple_df2 = mutate(simple_df, grade = ifelse(grade > 60, TRUE, FALSE))
3. If-else_if-else
-```
+```
simple_df3 = simple_df
simple_df3$grade = case_when(simple_df3$grade >= 90 ~ "A",
@@ -211,8 +211,10 @@ simple_df3$grade = case_when(simple_df3$grade >= 90 ~ "A",
or
-```
-simple_df3 = mutate(simple_df, grade = case_when(grade >= 90 ~ "A",
+```
+simple_df3 = simple_df
+
+simple_df3 = mutate(simple_df3, grade = case_when(grade >= 90 ~ "A",
grade >= 80 ~ "B",
grade >= 70 ~ "C",
grade >= 60 ~ "D",
@@ -244,7 +246,7 @@ if(expression_is_TRUE) {
3. If-else_if-else:
```
-if(expression_A_is_TRUE)
+if(expression_A_is_TRUE) {
#code goes here
}else if(expression_B_is_TRUE) {
#other code goes here
@@ -299,3 +301,7 @@ result
```
## [1] 5
```
+
+## Exercises
+
+You can find [exercises and solutions on Posit Cloud](https://posit.cloud/content/8236252), or on [GitHub](https://github.com/fhdsl/Intermediate_R_Exercises).
diff --git a/docs/no_toc/03-Data_cleaning_2.md b/docs/no_toc/03-Data_cleaning_2.md
index e2fd999..77397cf 100644
--- a/docs/no_toc/03-Data_cleaning_2.md
+++ b/docs/no_toc/03-Data_cleaning_2.md
@@ -1,14 +1,13 @@
# Data Cleaning, Part 2
-
```r
library(tidyverse)
```
## Tidy Data
-It is important to have standard of organizing data, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of **Tidy data**, developed by Hadley Wickham:
+It is important to have standard of organizing data, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The [principles of **Tidy data**](https://vita.had.co.nz/papers/tidy-data.html), developed by Hadley Wickham:
1. Each variable must have its own column.
@@ -221,7 +220,7 @@ ggplot(df) + aes(x = Q1_Sales, y = Q2_Sales, color = Store) + geom_point()
## Subjectivity in Tidy Data
-We have looked at clear cases of when a dataset is Tidy. In reality, the Tidy state depends on what we call variables and observations.
+We have looked at clear cases of when a dataset is Tidy. In reality, the Tidy state depends on what we call variables and observations. Consider this example, inspired by the following [blog post](https://kiwidamien.github.io/what-is-tidy-data.html) by Damien Martin.
```r
@@ -316,8 +315,6 @@ ggplot(kidney_long_still) + aes(x = treatment, y = recovery_rate, fill = stone_s
-## References
-
-https://vita.had.co.nz/papers/tidy-data.html
+## Exercises
-https://kiwidamien.github.io/what-is-tidy-data.html
+You can find [exercises and solutions on Posit Cloud](https://posit.cloud/content/8236252), or on [GitHub](https://github.com/fhdsl/Intermediate_R_Exercises).
diff --git a/docs/no_toc/04-Functions.md b/docs/no_toc/04-Functions.md
index f04bc37..dfa9c92 100644
--- a/docs/no_toc/04-Functions.md
+++ b/docs/no_toc/04-Functions.md
@@ -14,7 +14,7 @@ Some advice on writing functions:
- A function should do only one, well-defined task.
-### Anatomy of a function definition
+## Anatomy of a function definition
*Function definition consists of assigning a **function name** with a "function" statement that has a comma-separated list of named **function arguments**, and a **return expression**. The function name is stored as a variable in the global environment.*
@@ -34,13 +34,13 @@ With function definitions, not all code runs from top to bottom. The first four
When the function is called in line 5, the variables for the arguments are reassigned to function arguments to be used within the function and helps with the modular form. We need to introduce the concept of local and global environments to distinguish variables used only for a function from variables used for the entire program.
-### Local and global environments
+## Local and global environments
*{ } represents variable scoping: within each { }, if variables are defined, they are stored in a **local environment**, and is only accessible within { }. All function arguments are stored in the local environment. The overall environment of the program is called the **global environment** and can be also accessed within { }.*
The reason of having some of this "privacy" in the local environment is to make functions modular - they are independent little tools that should not interact with the rest of the global environment. Imagine someone writing a tool that they want to give someone else to use, but the tool depends on your environment, vice versa.
-### A step-by-step example
+## A step-by-step example
Using the `addFunction` function, let's see step-by-step how the R interpreter understands our code:
@@ -52,7 +52,7 @@ Using the `addFunction` function, let's see step-by-step how the R interpreter u

-### Function arguments create modularity
+## Function arguments create modularity
First time writers of functions might ask: why are variables we use for the arguments of a function *reassigned* for function arguments in the local environment? Here is an example when that process is skipped - what are the consequences?
@@ -81,7 +81,7 @@ Here is the execution for `w`:
The function did not work as expected because we used hard-coded variables from the global environment and not function argument variables unique to the function use!
-### Exercises
+## Examples
- Create a function, called `add_and_raise_power` in which the function takes in 3 numeric arguments. The function computes the following: the first two arguments are added together and raised to a power determined by the 3rd argument. The function returns the resulting value. Here is a use case: `add_and_raise_power(1, 2, 3) = 27` because the function will return this expression: `(1 + 2) ^ 3`. Another use case: `add_and_raise_power(3, 1, 2) = 16` because of the expression `(3 + 1) ^ 2`. Confirm with that these use cases work. Can this function used for numeric vectors?
@@ -114,7 +114,16 @@ The function did not work as expected because we used hard-coded variables from
## [1] 344 8
```
-- Create a function, called `medicaid_eligible` in which the function takes in one argument: a numeric vector called `age`. The function returns a numeric vector with the same length as `age`, in which elements are `0` for indicies that are less than 65 in `age`, and `1` for indicies 65 or higher in `age`. Use cases: `medicaid_eligible(c(30, 70)) = c(0, 1)`
+- Create a function, called `num_na` in which the function takes in any vector, and then return a single numeric value. This numeric value is the number of `NA`s in the vector. Use cases: `num_na(c(NA, 2, 3, 4, NA, 5)) = 2` and `num_na(c(2, 3, 4, 5)) = 0`. Hint 1: Use `is.na()` function. Hint 2: Given a logical vector, you can count the number of `TRUE` values by using `sum()`, such as `sum(c(TRUE, TRUE, FALSE)) = 2`.
+
+
+ ```r
+ num_na = function(x) {
+ return(sum(is.na(num_na)))
+ }
+ ```
+
+- Create a function, called `medicaid_eligible` in which the function takes in one argument: a numeric vector called `age`. The function returns a numeric vector with the same length as `age`, in which elements are `0` for indicies that are less than 65 in `age`, and `1` for indicies 65 or higher in `age`. (Hint: This is a data recoding problem!) Use cases: `medicaid_eligible(c(30, 70)) = c(0, 1)`
```r
@@ -130,3 +139,7 @@ The function did not work as expected because we used hard-coded variables from
```
## [1] 0 1
```
+
+## Exercises
+
+You can find [exercises and solutions on Posit Cloud](https://posit.cloud/content/8236252), or on [GitHub](https://github.com/fhdsl/Intermediate_R_Exercises).
diff --git a/docs/no_toc/05-Iteration.md b/docs/no_toc/05-Iteration.md
new file mode 100644
index 0000000..6effb67
--- /dev/null
+++ b/docs/no_toc/05-Iteration.md
@@ -0,0 +1,324 @@
+# Iteration
+
+Suppose that you want to repeat a chunk of code many times, but changing one variable's value each time you do it. This could be modifying each element of a vector with the same operation, or analyzing a dataframe with different parameters.
+
+There are three common strategies to go about this:
+
+1. Copy and paste the code chunk, and change that variable's value. Repeat. *This can be a starting point in your analysis, but will lead to errors easily.*
+2. Use a `for` loop to repeat the chunk of code, and let it loop over the changing variable's value. *This is popular for many programming languages, but the R programming culture encourages a functional way instead*.
+3. **Functionals** allow you to take a function that solves the problem for a single input and generalize it to handle any number of inputs. *This is very popular in R programming culture.*
+
+## For loops
+
+A `for` loop repeats a chunk of code many times, once for each element of an input vector.
+
+```
+for (my_element in my_vector) {
+ chunk of code
+}
+```
+
+Most often, the "chunk of code" will make use of `my_element`.
+
+#### We can loop through the indicies of a vector
+
+The function `seq_along()` creates the indicies of a vector. It has almost the same properties as `1:length(my_vector)`, but avoids issues when the vector length is 0.
+
+
+```r
+my_vector = c(1, 3, 5, 7)
+
+for(i in seq_along(my_vector)) {
+ print(my_vector[i])
+}
+```
+
+```
+## [1] 1
+## [1] 3
+## [1] 5
+## [1] 7
+```
+
+#### Alternatively, we can loop through the elements of a vector
+
+
+```r
+for(vec_i in my_vector) {
+ print(vec_i)
+}
+```
+
+```
+## [1] 1
+## [1] 3
+## [1] 5
+## [1] 7
+```
+
+#### Another example via indicies
+
+
+```r
+result = rep(NA, length(my_vector))
+for(i in seq_along(my_vector)) {
+ result[i] = log(my_vector[i])
+}
+```
+
+## Functionals
+
+A **functional** is a function that takes in a data structure and function as inputs and applies the function on the data structure, element by element. It *maps* your input data structure to an output data structure based on the function. It encourages the usage of modular functions in your code.
+
+
+
+Or,
+
+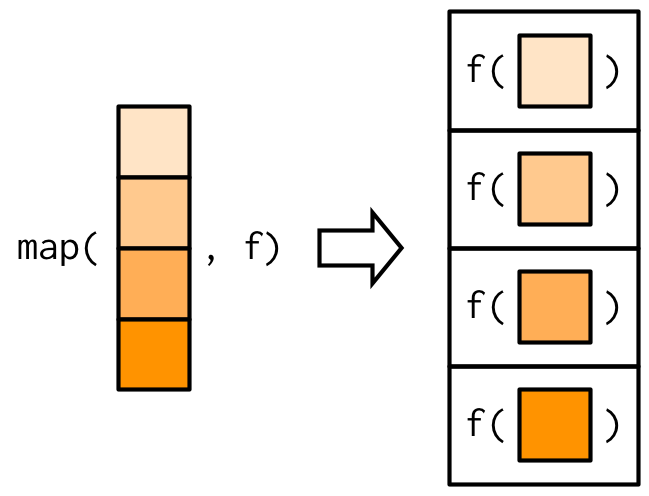{width="250"}
+
+We will use the `purrr` package in `tidyverse` to use functionals.
+
+`map()` takes in a vector or a list, and then applies the function on each element of it. The output is *always* a list.
+
+
+
+
+```r
+my_vector = c(1, 3, 5, 7)
+map(my_vector, log)
+```
+
+```
+## [[1]]
+## [1] 0
+##
+## [[2]]
+## [1] 1.098612
+##
+## [[3]]
+## [1] 1.609438
+##
+## [[4]]
+## [1] 1.94591
+```
+
+Lists are useful if what you are using it on requires a flexible data structure.
+
+To be more specific about the output type, you can do this via the `map_*` function, where `*` specifies the output type: `map_lgl()`, `map_chr()`, and `map_dbl()` functions return vectors of logical values, strings, or numbers respectively.
+
+For example, to make sure your output is a double (numeric):
+
+
+```r
+map_dbl(my_vector, log)
+```
+
+```
+## [1] 0.000000 1.098612 1.609438 1.945910
+```
+
+All of these are toy examples that gets us familiar with the syntax, but we already have built-in functions to solve these problems, such as `log(my_vector)`. Let's see some real-life case studies.
+
+## Case studies
+
+### 1. Loading in multiple files.
+
+Suppose that we want to load in a few dataframes, and store them in a list of dataframes for analysis downstream.
+
+We start with the filepaths we want to load in as dataframes.
+
+
+```r
+paths = c("classroom_data/students.csv", "classroom_data/CCLE_metadata.csv")
+```
+
+The function we want to use to load the data in will be `read_csv()`.
+
+Let's practice writing out one iteration:
+
+
+```r
+result = read_csv(paths[1])
+```
+
+#### To do this functionally, we think about:
+
+- What variable we need to loop through: `paths`
+
+- The repeated task as a function: `read_csv()`
+
+- The looping mechanism, and its output: `map()` outputs lists.
+
+
+```r
+loaded_dfs = map(paths, read_csv)
+```
+
+#### To do this with a for loop, we think about:
+
+- What variable we need to loop through: `paths`.
+
+- Do we need to store the outcome of this loop in a data structure? Yes, a list.
+
+- At each iteration, what are we doing? Use `read_csv()` on the current element, and store it in the output list.
+
+
+```r
+paths = c("classroom_data/students.csv", "classroom_data/CCLE_metadata.csv")
+loaded_dfs = vector(mode = "list", length = length(paths))
+for(i in seq_along(paths)) {
+ df = read_csv(paths[i])
+ loaded_dfs[[i]] = df
+}
+```
+
+### 2. Analyze a dataframe with different parameters.
+
+Suppose you are working with the `penguins` dataframe:
+
+
+```r
+library(palmerpenguins)
+head(penguins)
+```
+
+```
+## # A tibble: 6 × 8
+## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
+##
+## 1 Adelie Torgersen 39.1 18.7 181 3750
+## 2 Adelie Torgersen 39.5 17.4 186 3800
+## 3 Adelie Torgersen 40.3 18 195 3250
+## 4 Adelie Torgersen NA NA NA NA
+## 5 Adelie Torgersen 36.7 19.3 193 3450
+## 6 Adelie Torgersen 39.3 20.6 190 3650
+## # ℹ 2 more variables: sex , year
+```
+
+and you want to look at the mean `bill_length_mm` for each of the three species (Adelie, Chinstrap, Gentoo).
+
+Let's practice writing out one iteration:
+
+
+```r
+species_to_analyze = c("Adelie", "Chinstrap", "Gentoo")
+penguins_subset = filter(penguins, species == species_to_analyze[1])
+mean(penguins_subset$bill_length_mm, na.rm = TRUE)
+```
+
+```
+## [1] 38.79139
+```
+
+#### To do this functionally, we think about:
+
+- What variable we need to loop through: `c("Adelie", "Chinstrap", "Gentoo")`
+
+- The repeated task as a function: a custom function that takes in a specie of interest. The function filters the rows of `penguins` to the species of interest, and compute the mean of `bill_length_mm`.
+
+- The looping mechanism, and its output: `map_dbl()` outputs (double) numeric vectors.
+
+
+```r
+analysis = function(current_species) {
+ penguins_subset = dplyr::filter(penguins, species == current_species)
+ return(mean(penguins_subset$bill_length_mm, na.rm=TRUE))
+}
+
+map_dbl(c("Adelie", "Chinstrap", "Gentoo"), analysis)
+```
+
+```
+## [1] 38.79139 48.83382 47.50488
+```
+
+#### To do this with a for loop, we think about:
+
+- What variable we need to loop through: `c("Adelie", "Chinstrap", "Gentoo")`.
+
+- Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector.
+
+- At each iteration, what are we doing? Filter the rows of `penguins` to the species of interest, and compute the mean of `bill_length_mm`.
+
+
+```r
+outcome = rep(NA, length(species_to_analyze))
+for(i in seq_along(species_to_analyze)) {
+ penguins_subset = filter(penguins, species == species_to_analyze[i])
+ outcome[i] = mean(penguins_subset$bill_length_mm, na.rm=TRUE)
+}
+outcome
+```
+
+```
+## [1] 38.79139 48.83382 47.50488
+```
+
+### 3. Calculate summary statistics on columns of a dataframe.
+
+Suppose that you are interested in the numeric columns of the `penguins` dataframe.
+
+
+```r
+penguins_numeric = penguins %>% select(bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g)
+```
+
+and you are interested to look at the mean of each column. It is very helpful to interpret the dataframe `penguins_numeric` as a *list*, iterating through each column as an element of a list.
+
+Let's practice writing out one iteration:
+
+
+```r
+mean(penguins_numeric[[1]], na.rm = TRUE)
+```
+
+```
+## [1] 43.92193
+```
+
+#### To do this functionally, we think about:
+
+- What variable we need to loop through: the list `penguins_numeric`
+
+- The repeated task as a function: `mean()` with the argument `na.rm = TRUE`.
+
+- The looping mechanism, and its output: `map_dbl()` outputs (double) numeric vectors.
+
+
+```r
+map_dbl(penguins_numeric, mean, na.rm = TRUE)
+```
+
+```
+## bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
+## 43.92193 17.15117 200.91520 4201.75439
+```
+
+Here, R is interpreting the dataframe `penguins_numeric` as a *list*, iterating through each column as an element of a list:
+
+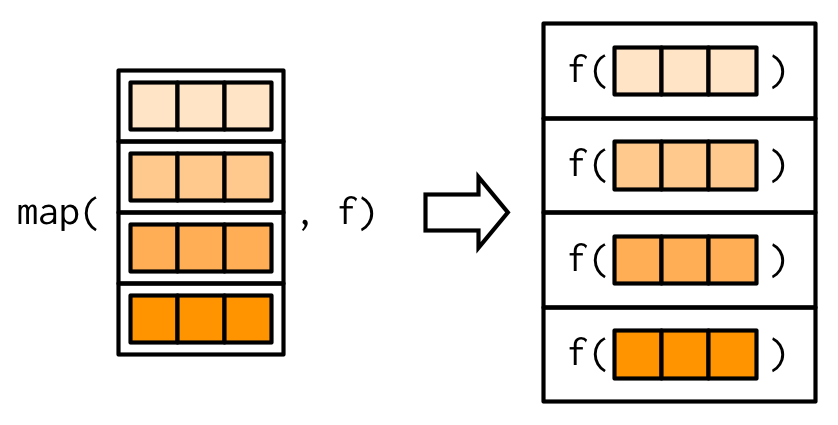{width="300"}
+
+#### To do this with a for loop, we think about:
+
+- What variable we need to loop through: the elements of `penguins_numeric` as a list.
+
+- Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector.
+
+- At each iteration, what are we doing? Compute the mean of an element of `penguins_numeric`.
+
+
+```r
+result = rep(NA, ncol(penguins_numeric))
+for(i in seq_along(penguins_numeric)) {
+ result[i] = mean(penguins_numeric[[i]], na.rm = TRUE)
+}
+result
+```
+
+```
+## [1] 43.92193 17.15117 200.91520 4201.75439
+```
+
+## Exercises
+
+You can find [exercises and solutions on Posit Cloud](https://posit.cloud/content/8236252), or on [GitHub](https://github.com/fhdsl/Intermediate_R_Exercises).
diff --git a/docs/no_toc/404.html b/docs/no_toc/404.html
index 4da1576..41eb66e 100644
--- a/docs/no_toc/404.html
+++ b/docs/no_toc/404.html
@@ -138,6 +138,7 @@
5.1 Part 1: Looking at documentation to load in data
-
Suppose that you want to load in data “students.csv” in a CSV format, and you don’t know what tools to use. You decide to see whether the package “readr” can be useful to solve your problem. Where should you look?
-
All R packages must be stored on CRAN (Comprehensive R Archive Network), and all packages have a website that points to the reference manual (what is pulled up using the ? command), source code, vignettes examples, and dependencies on other packages. Here is the website for “readr”.
-
In the package, you find some potential functions for importing your data:
-
-
read_csv("file.csv") for comma-separated files
-
read_tsv("file.tsv") for tab-deliminated files
-
read_excel("example.xlsx") for excel files
-
read_excel("example.xlsx", sheet = "sheet1") for excel files with a sheet name
-
read_delim() for general-deliminated files, such as: read_delim("file.csv", sep = ",").
-
-
After looking at the vignettes, it seems that read_csv() is a function to try.
-
Let’s look at the read_csv() function documentation, which can be accessed via ?read_csv.
We see that the only required argument is the file variable, which is documented to be “Either a path to a file, a connection, or literal data (either a single string or a raw vector).” All the other arguments are considered optional, because they have a pre-allocated value in the documentation.
-
Load in “students.csv” via read_csv() function as a dataframe variable students and take a look at its contents via View().
-
library(tidyverse)
-
## Warning: package 'tidyverse' was built under R version 4.0.3
Something looks weird here. There is only one column, and it seems that the first two entries start with “#”, and don’t fit a CSV file format. These first two entries that start with “#” likely are comments giving metadata about the file, and they should be ignore when loading in the data.
-
Let’s try again. Take a look at the documentation for the comment argument and give it a character value "#" with this situation. Any text after the comment characters will be silently ignored.
-
The column names are not very consistent . Take a look at the documentation for the col_names argument and give it a value of c("student_id", "full_name", "favorite_food", "meal_plan", "age").
-
Alternatively, you could have loaded the data in without col_names option and modified the column names by accessing names(students).
-
For more information on loading in data, see this chapter of R for Data Science.
-
-
-
5.2 Part 2: Recoding data: warm-up
-
Consider this vector:
-
scores =c(23, 46, -3, 5, -1)
-
Recode scores so that all the negative values are 0.
-
Let’s look at the values of students dataframe more carefully. We will do some recoding on this small dataframe. It may feel trivial because you could do this by hand in Excel, but this is a practice on how we can scale this up with larger datasets!
-
Notice that some of the elements of this dataframe has proper NA values and also a character “N/A”. We want “N/A” to be a proper NA value.
-
Recode “N/A” to NA in the favorite_food column:
-
Recode “five” to 5 in the age column:
-
Create a new column age_category so that it has value “toddler” if age is < 6, and “child” if age is >= 6.
-
(Hint: You can create a new column via mutate, or you can directly refer to the new column via student$``age_category.)
-
Create a new column favorite_food_numeric so that it has value 1 if favorite_food is “Breakfast and lunch”, 2 if “Lunch only”, and 3 if “Dinner only”.
-
-
-
5.3 Part 3: Recoding data in State Cancer Profiles
[State Cancer Profile data] was developed with the idea to provide a geographic profile of cancer burden in the United States and reveal geographic disparities in cancer incidence, mortality, risk factors for cancer, and cancer screening, across different population subgroups.
-
-
In this analysis, we want to examine cancer incidence rates in state of Washington and make some comparisons between groups. The cancer incidence rate can be accessed at this website, once you give input of what kind of incidence data you want to access. If you want to analyze this data in R, it takes a bit of work of exporting the data and loading it into R.
-
To access this data easier in R, DaSL staff built a R package cancerprof to easily load in the data. Let’s look at the package’s documentation to see how to get access to cancer incidence data.
-
Towards the bottom of the documentation are some useful examples to consider as starting point.
-
Load in data about the following population: melanoma incidence in WA at the county level for males of all ages, all cancer stages, averaged in the past 5 years. Store it as a dataframe variable named melanoma_incidence
-
(If you are stuck, you can use the first example in the documentation.)
-
Take a look at the data yourself and explore it.
-
Let’s select a few columns of interest and give them column names that doesn’t contain spaces. We can access column names with spaces via the backtick ` symbol.
-
#uncomment to run!
-
-#melanoma_incidence = select(melanoma_incidence, County, `Age Adjusted Incidence Rate`, `Recent Trend`)
-
-#names(melanoma_incidence) = c("County", "Age_adjusted_incidence_rate", "Recent_trend")
-
Take a look at the column Age_adjusted_incidence_rate. It has missing data coded as “*” (notice the space after *). Recode “*” as NA.
-
Finally, notice that the data type for Age_adjusted_incidence_rate is character, if you run the function is.character() or class() on it. Convert it to a numeric data type.
As you become more independent R programmers, you will spend time learning about new functions on your own. We have gone over the basic anatomy of a function call back in Intro to R, but now let’s go a bit deeper to understand how a function is built and how to call them.
Recall that a function has a function name, input arguments, and a return value.
Function definition consists of assigning a function name with a “function” statement that has a comma-separated list of named function arguments, and a return expression. The function name is stored as a variable in the global environment.
@@ -309,8 +297,8 @@
4.1 Interpreting functions, caref
...: further arguments passed to or from other methods.
Notice that the arguments trim = 0, na.rm = FALSE have default values. This means that these arguments are optional - you should provide it only if you want to. With this understanding, you can use mean() in a new way:
The use of . . . (dot-dot-dot): This is a special argument that allows a function to take any number of arguments. This isn’t very useful for the mean() function, but it makes sense for function such as select() and filter(), and mutate(). For instance, in select(), once you provide your dataframe for the argument .data, you can pile on as many columns to select in the rest of the argument.
Usage:
@@ -329,8 +317,8 @@
4.1 Interpreting functions, caref
select a range of variables.
You will look at the function documentation on your own to see how to deal with more complex cases.
-
-
4.2 Recoding Data / Conditionals
+
+
3.2 Recoding Data / Conditionals
It is often said that 80% of data analysis is spent on the cleaning and preparing data. Today we will start looking at common data cleaning tasks. Suppose that you have a column in your data that needs to be recoded. Since a dataframe’s column, when selected via $, is a vector, let’s start talking about recoding vectors. If we have a numeric vector, then maybe you want to have certain values to be out of bounds, or assign a range of values to a character category. If we have a character vector, then maybe you want to reassign it to a different value.
Here are popular recoding logical scenarios:
@@ -339,17 +327,17 @@
4.2 Recoding Data / Conditionals<
If-else_if-else: “If elements of the vector meets condition A, then they are assigned value X. Else, if the elements of the vector meets condition B, they are assigned value Y. Otherwise, they are assigned value Z.”
Let’s look at a vector of grade values, as an example:
-
grade =c(90, 78, 95, 74, 56, 81, 102)
+
grade =c(90, 78, 95, 74, 56, 81, 102)
If
Instead of having the bracket [ ] notation on the right hand side of the equation, if it is on the left hand side of the equation, then we can modify a subset of the vector.
The 3 common scenarios we looked at for recoding data is closely tied to the concept of conditionals in programming: given certain conditions, you run a specific code chunk. Given a vector’s value, assign it a different value. Or, given a value, run the following hundred lines of code. Here is what it looks like:
If:
@@ -410,7 +400,7 @@
4.3 Conditionals
If-else_if-else:
-
if(expression_A_is_TRUE)
+
if(expression_A_is_TRUE) {
#code goes here
}else if(expression_B_is_TRUE) {
#other code goes here
@@ -419,34 +409,38 @@
4.3 Conditionals
}
The expression that is being tested whether it is TRUEmust be a singular logical value, and not a logical vector. If the latter, see the recoding section for now.
It is important to have standard of organizing data, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of Tidy data, developed by Hadley Wickham:
+
+
Chapter 4 Data Cleaning, Part 2
+
library(tidyverse)
+
+
4.1 Tidy Data
+
It is important to have standard of organizing data, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of Tidy data, developed by Hadley Wickham:
Each variable must have its own column.
Each observation must have its own row.
@@ -274,22 +262,22 @@
6.1 Tidy Data
Multiple variables are stored in a single column
After some clear examples, we emphasize that “Tidy” data is subjective to what kind of analysis you want to do with the dataframe.
-
-
6.1.1 1. Columns contain values, rather than variables (Long is tidy)
## Store Year Q1_Sales Q2_Sales Q3_Sales Q4_Sales
## 1 A 2018 55 45 22 50
## 2 B 2018 98 70 60 60
Each observation is a store, and each observation has its own row. That looks good.
The columns “Q1_Sales”, …, “Q4_Sales” seem to be values of a single variable “quarter” of our observation. The values of “quarter” are not in a single column, but are instead in the columns.
## # A tibble: 8 × 4
## Store Year quarter sales
## <chr> <dbl> <chr> <dbl>
@@ -305,20 +293,20 @@
6.1.1 1. Columns contain values,
The new columns “quarter” and “sales” are variables that describes our observation, and describes our values. We’re in a tidy state!
We have transformed our data to a “longer” format, as our observation represents something more granular or detailed than before. Often, the original variables values will repeat itself in a “longer format”. We call the previous state of our dataframe is a “wider” format.
-
-
6.1.2 2. Variables are stored in rows (Wide is tidy)
+
+
4.1.2 2. Variables are stored in rows (Wide is tidy)
Are all tidy dataframes Tidy in a “longer” format?
6.1.2 2. Variables are stored in
## 4 B KRAS_expression 3.9
Here, each observation is a sample’s gene…type? The observation feels awkward because variables are stored in rows. Also, the column “values” contains multiple variable types: gene expression and mutation values that got coerced to numeric!
We are back to our orignal form, and it was already Tidy.
-
-
6.1.3 3. Multiple variables are stored in a single column
-
table3
+
+
4.1.3 3. Multiple variables are stored in a single column
+
table3
## # A tibble: 6 × 3
## country year rate
## * <chr> <int> <chr>
@@ -351,7 +339,7 @@
6.1.3 3. Multiple variables are s
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583
There seems to be two variables in the numerator and denominator of “rate” column. Let’s separate it.
-
separate(table3, col ="rate", into =c("count", "population"), sep ="/")
+
separate(table3, col ="rate", into =c("count", "population"), sep ="/")
## # A tibble: 6 × 4
## country year count population
## <chr> <int> <chr> <chr>
@@ -363,10 +351,10 @@
6.1.3 3. Multiple variables are s
## 6 China 2000 213766 1280428583
-
-
6.2 Uses of Tidy data
+
+
4.2 Uses of Tidy data
In general, many functions for analysis and visualization in R assumes that the input dataframe is Tidy. These tools assumes the values of each variable fall in their own column vector. For instance, from our first example, we can compare sales across quarters and stores.
-
df_long
+
df_long
## # A tibble: 8 × 4
## Store Year quarter sales
## <chr> <dbl> <chr> <dbl>
@@ -378,21 +366,21 @@
6.2 Uses of Tidy data
## 6 B 2018 Q2_Sales 70
## 7 B 2018 Q3_Sales 60
## 8 B 2018 Q4_Sales 60
-
ggplot(df_long) +aes(x = quarter, y = sales, group = Store) +geom_point() +geom_line()
+
ggplot(df_long) +aes(x = quarter, y = sales, group = Store) +geom_point() +geom_line()
Although in its original state we can also look at sales between quarter, we can only look between two quarters at once. Tidy data encourages looking at data in the most granular scale.
-
ggplot(df) +aes(x = Q1_Sales, y = Q2_Sales, color = Store) +geom_point()
+
ggplot(df) +aes(x = Q1_Sales, y = Q2_Sales, color = Store) +geom_point()
-
-
6.3 Subjectivity in Tidy Data
-
We have looked at clear cases of when a dataset is Tidy. In reality, the Tidy state depends on what we call variables and observations.
We have looked at clear cases of when a dataset is Tidy. In reality, the Tidy state depends on what we call variables and observations. Consider this example, inspired by the following blog post by Damien Martin.
Right now, the kidney dataframe clearly has values of a variable in the column. Let’s try to make it Tidy by making it into a longer form and separating out variables that are together in a column.
The reason why both of these versions seem Tidy is that the columns “recovered” and “failed” can be interpreted as independent variables and values of the variable “treatment”.
Ultimately, we decide which dataframe we prefer based on the analysis we want to do.
For instance, when our observation is about a kidney stone’s treatment’s outcome type, we compare it between outcome type, treatment, and stone size.
-
ggplot(kidney_long) +aes(x = treatment, y = count, fill = outcome) +geom_bar(position="dodge", stat="identity") +facet_wrap(~stone_size)
+
ggplot(kidney_long) +aes(x = treatment, y = count, fill = outcome) +geom_bar(position="dodge", stat="identity") +facet_wrap(~stone_size)
When our observation is about a kidney stone’s treatment’s, we compare a new variable recovery rate ( = recovered / (recovered + failed)) between treatment and stone size.
How do you subset the following vector to the first three elements?
-
measurements =c(2, 4, -1, -3, 2, -1, 10)
-
How do you subset the original vector so that it only has negative values?
-
How do you subset the following vector so that it has no NA values?
-
vec_with_NA =c(2, 4, NA, NA, 3, NA)
-
Consider the following logical vector some_logicals. Convert Logical vector -> Numeric vector -> Character vector in two steps. Check that you are doing this correctly along the way by using the class() function, or is.numeric() and is.character(), on the converted data.
patient =list(
-name =" ",
-age =34,
-pronouns =c("he", "him", "/", "they", "them"),
-vaccines =c("hep-B", "chickenpox", "HPV"),
-visits =NA
-)
-
-visit1 =list(
-symptoms =c("runny nose", "sore throat", "frustration"),
-prescription ="recommended time off from work, rest.",
-date ="1/1/2000"
-)
-
-visit2 =list(
-symptoms =c("fainted", "pale complexion"),
-prescription ="drink water and take time off work.",
-date ="1/1/2001"
-)
-
Access the first element of patient via double brackets [[ ]] and modify it to a value of your choice.
-
Access the named element “pronouns” of patient via double bracket [[ ]] or $ and modify its value so that it doesn’t contain the “/” element. (Use your vector subsetting skills here after you access the appropriate element from the list.)
-
Create a new list all_visits with elements visit1 and visit2. Yes, we’re making lists within lists!
-
Suppose you want to use all_visits to access visit 1’s symptoms. You would continue the double brackets [[ ]] or $ notation: all_visits[[1]] returns a list, so we access the first element of that list via all_visits[[1]][[1]].
How would you use all_visits to access visit 2’s prescription?
-
How would you use all_visits to access visit 2’s symptom element “pale complexion”? Remember, once you access a vector, you would go back to the single bracket [ ] to access its elements.
-
Finally, assign all_visits to patient’s visits.
-
-
3.2.1 Part 3: Dataframes (Lists)
-
A dataframe is just a named list of vectors of same length with attributes of (column) names and row.names.
-
library(palmerpenguins)
-head(penguins)
-
## # A tibble: 6 × 8
-## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
-## <fct> <fct> <dbl> <dbl> <int> <int>
-## 1 Adelie Torgersen 39.1 18.7 181 3750
-## 2 Adelie Torgersen 39.5 17.4 186 3800
-## 3 Adelie Torgersen 40.3 18 195 3250
-## 4 Adelie Torgersen NA NA NA NA
-## 5 Adelie Torgersen 36.7 19.3 193 3450
-## 6 Adelie Torgersen 39.3 20.6 190 3650
-## # ℹ 2 more variables: sex <fct>, year <int>
-
Access the body_mass_g column vector of penguins via the double bracket [[ ]], treating penguins like a list, and compute the mean. Remember to use na.rm = TRUE to remove any NA values: mean(x, na.rm = TRUE)
-
Create a new dataframe penguins_clean, which has no NA values in the body_mass_g column. You need to filter out rows that have NAs in the column bill_length_mm:
-
Now, subset penguins_clean to each of the three species and compute their respective mean value of body_mass_g. Because you already got rid of NAs in body_mass_g, you can just use mean(x) without the extra option. How do they compare?
-
Finally, make a box plot of species (x-axis) vs. body_mass_g (y-axis) via penguins_clean dataframe. I’ll get you started…
-
#ggplot(penguins_clean) + aes(x = , y = ) + geom_boxplot()
Continue building programming fundamentals: how to make use of complex data structures, use custom functions built by other R users, and creating your own functions. How to iterate repeated tasks that scales naturally.
+
Continue building programming fundamentals: How to use complex data structures, use and create custom functions, and how to iterate repeated tasks
Continue exploration of data science fundamentals: how to clean messy data to a Tidy form for analysis.
-
Outcome: Conduct a full analysis in the data science workflow (minus model).
+
At the end of the course, you will be able to: conduct a full analysis in the data science workflow (minus model).
@@ -482,7 +470,7 @@
2.7 Lists
l1$score
## [1] 2.3 5.9
Therefore, l1$score is the same as l1[[4]] and is the same as l1[["score"]].
-
A dataframe is just a named list of vectors of same length with attributes of (column) names and row.names.
+
A dataframe is just a named list of vectors of same length with additional attributes of (column) names and row.names.
The course is intended for researchers who want to continue learning the fundamentals of R programming and how to deal with messy datasets. The audience should know how to subset dataframes and vectors and conduct basic analysis, and/or have taken our Intro to R course.
+
+
+
1.3 Offerings
+
This course is taught on a regular basis at Fred Hutch Cancer Center through the Data Science Lab. Announcements of course offering can be found here. If you wish to follow the course content asynchronously, you may access the course content on this website and exercises and solutions on Posit Cloud. The Posit Cloud compute space can be copied to your own workspace for personal use, or you can access the exercises and solutions on GitHub.
diff --git a/docs/no_toc/index.md b/docs/no_toc/index.md
index e427fbe..0aae292 100644
--- a/docs/no_toc/index.md
+++ b/docs/no_toc/index.md
@@ -1,6 +1,6 @@
---
title: "Intermediate R, Season 3"
-date: "March, 2024"
+date: "May, 2024"
site: bookdown::bookdown_site
documentclass: book
bibliography: [book.bib]
@@ -15,11 +15,14 @@ output:
# About this Course
-## Curriculum
+## Curriculum
-The course continues building programming fundamentals in R programming and data analysis. You will learn how to make use of complex data structures, use custom functions built by other R users, creating your own functions, and how to iterate repeated tasks that scales naturally. You will also learn how to clean messy data to a Tidy form for analysis, and conduct an end-to-end data science workflow.
+The course continues building programming fundamentals in R programming and data analysis. You will learn how to make use of complex data structures, use custom functions built by other R users, creating your own functions, and how to iterate repeated tasks that scales naturally. You will also learn how to clean messy data to a Tidy form for analysis, and conduct an end-to-end data science workflow.
-## Target Audience
+## Target Audience
-The course is intended for researchers who want to continue learning the fundamentals of R programming and how to deal with messy datasets. The audience should know how to subset dataframes and vectors and conduct basic analysis, and/or have taken our [Intro to R course](https://github.com/fhdsl/Intro_to_R).
+The course is intended for researchers who want to continue learning the fundamentals of R programming and how to deal with messy datasets. The audience should know how to subset dataframes and vectors and conduct basic analysis, and/or have taken our [Intro to R course](https://github.com/fhdsl/Intro_to_R).
+## Offerings
+
+This course is taught on a regular basis at Fred Hutch Cancer Center through the Data Science Lab. Announcements of course offering can be found [here](https://hutchdatascience.org/training/). If you wish to follow the course content asynchronously, you may access the course content on this website and [exercises and solutions on Posit Cloud](https://posit.cloud/content/8236252). The Posit Cloud compute space can be copied to your own workspace for personal use, or you can access the [exercises and solutions on GitHub](https://github.com/fhdsl/Intermediate_R_Exercises).
diff --git a/docs/no_toc/repeating-tasks.html b/docs/no_toc/iteration.html
similarity index 51%
rename from docs/no_toc/repeating-tasks.html
rename to docs/no_toc/iteration.html
index b0afc8b..8294898 100644
--- a/docs/no_toc/repeating-tasks.html
+++ b/docs/no_toc/iteration.html
@@ -4,11 +4,11 @@
- Chapter 10 Repeating tasks | Intermediate R, Season 3
-
+ Chapter 6 Iteration | Intermediate R, Season 3
+
-
+
@@ -16,7 +16,7 @@
-
+
@@ -29,8 +29,8 @@
-
-
+
+
@@ -138,6 +138,7 @@
Suppose that you want to repeat a chunk of code many times, but changing one variable’s value each time you do it. This could be modifying each element of a vector with the same operation, or analyzing a dataframe with different parameters.
There are three common strategies to go about this:
@@ -256,106 +244,111 @@
Chapter 10 Repeating tasks
Use a for loop to repeat the chunk of code, and let it loop over the changing variable’s value. This is popular for many programming languages, but the R programming culture encourages a functional way instead.
Functionals allow you to take a function that solves the problem for a single input and generalize it to handle any number of inputs. This is very popular in R programming culture.
-
-
10.1 For loops
+
+
6.1 For loops
A for loop repeats a chunk of code many times, once for each element of an input vector.
for (my_element in my_vector) {
chunk of code
}
Most often, the “chunk of code” will make use of my_element.
-
-
10.1.0.1 We can loop through elements of a vector and print it out.
6.1.0.1 We can loop through the indicies of a vector
+
The function seq_along() creates the indicies of a vector. It has almost the same properties as 1:length(my_vector), but avoids issues when the vector length is 0.
6.1.0.2 Alternatively, we can loop through the elements of a vector
+
for(vec_i in my_vector) {
+print(vec_i)
+}
## [1] 1
## [1] 3
## [1] 5
## [1] 7
+
+
6.1.0.3 Another example via indicies
+
result =rep(NA, length(my_vector))
+for(i inseq_along(my_vector)) {
+ result[i] =log(my_vector[i])
+}
+
-
-
10.2 Functionals
+
+
6.2 Functionals
A functional is a function that takes in a data structure and function as inputs and applies the function on the data structure, element by element. It maps your input data structure to an output data structure based on the function. It encourages the usage of modular functions in your code.
Or,
We will use the purrr package in tidyverse to use functionals.
-
map() takes in a vector or a list, and then applies the function on each element of it. The output is always a list. (You see the output twice, because it prints out element by element, and then returns a list.)
-
map(my_vector, print)
-
## [1] 1
-## [1] 3
-## [1] 5
-## [1] 7
+
map() takes in a vector or a list, and then applies the function on each element of it. The output is always a list.
Lists are useful if what you are using it on requires a flexible data structure.
To be more specific about the output type, you can do this via the map_* function, where * specifies the output type: map_lgl(), map_chr(), and map_dbl() functions return vectors of logical values, strings, or numbers respectively.
For example, to make sure your output is a double (numeric):
-
map_dbl(my_vector, log)
+
map_dbl(my_vector, log)
## [1] 0.000000 1.098612 1.609438 1.945910
-
All of these are toy examples that gets us familiar with the syntax, but we already have built-in functions to solve these problems. Let’s see some real-life case studies.
+
All of these are toy examples that gets us familiar with the syntax, but we already have built-in functions to solve these problems, such as log(my_vector). Let’s see some real-life case studies.
-
-
10.3 Case studies
-
-
10.3.1 1. Loading in multiple dataframes from files for analysis
+
+
6.3 Case studies
+
+
6.3.1 1. Loading in multiple files.
Suppose that we want to load in a few dataframes, and store them in a list of dataframes for analysis downstream.
We start with the filepaths we want to load in as dataframes.
What variable we need to loop through: c("Adelie", "Chinstrap", "Gentoo")
The repeated task as a function: a custom function that takes in a specie of interest. The function filters the rows of penguins to the species of interest, and compute the mean of bill_length_mm.
The looping mechanism, and its output: map_dbl() outputs (double) numeric vectors.
and you are interested to look at the mean of each column. It is very helpful to interpret the dataframe penguins_numeric as a list, iterating through each column as an element of a list.
Let’s practice writing out one iteration:
-
mean(penguins_numeric[[1]], na.rm =TRUE)
+
mean(penguins_numeric[[1]], na.rm =TRUE)
## [1] 43.92193
-
-
10.3.3.1 To do this functionally, we think about:
+
+
6.3.3.1 To do this functionally, we think about:
What variable we need to loop through: the list penguins_numeric
The repeated task as a function: mean() with the argument na.rm = TRUE.
The looping mechanism, and its output: map_dbl() outputs (double) numeric vectors.
-
-
-
-
-
-
-
-
-
-
-
-
-
diff --git a/docs/no_toc/search_index.json b/docs/no_toc/search_index.json
index c0fde75..46b5cf8 100644
--- a/docs/no_toc/search_index.json
+++ b/docs/no_toc/search_index.json
@@ -1 +1 @@
-[["index.html", "Intermediate R, Season 3 Chapter 1 About this Course 1.1 Curriculum 1.2 Target Audience", " Intermediate R, Season 3 March, 2024 Chapter 1 About this Course 1.1 Curriculum The course continues building programming fundamentals in R programming and data analysis. You will learn how to make use of complex data structures, use custom functions built by other R users, creating your own functions, and how to iterate repeated tasks that scales naturally. You will also learn how to clean messy data to a Tidy form for analysis, and conduct an end-to-end data science workflow. 1.2 Target Audience The course is intended for researchers who want to continue learning the fundamentals of R programming and how to deal with messy datasets. The audience should know how to subset dataframes and vectors and conduct basic analysis, and/or have taken our Intro to R course. "],["fundamentals.html", "Chapter 2 Fundamentals 2.1 Goals of this course 2.2 Data types in R 2.3 Data structures 2.4 Vector 2.5 Factors 2.6 Dataframes 2.7 Lists 2.8 Matrix", " Chapter 2 Fundamentals 2.1 Goals of this course Continue building programming fundamentals: how to make use of complex data structures, use custom functions built by other R users, and creating your own functions. How to iterate repeated tasks that scales naturally. Continue exploration of data science fundamentals: how to clean messy data to a Tidy form for analysis. Outcome: Conduct a full analysis in the data science workflow (minus model). 2.2 Data types in R Numeric: 18, -21, 65, 1.25 Character: “ATCG”, “Whatever”, “948-293-0000” Logical: TRUE, FALSE Missing values: NA 2.3 Data structures Vector Factor Dataframe List Matrix 2.4 Vector We know what an (atomic) vector is: it can contains a data type, and all elements must be the same data type. Within the Numeric type that we are familiar with, there are more specific types: Integer consists of whole number values, and Double consists of decimal values. Most of the time we only need to consider Numeric types, but once in a while we need to be more specific. We can test whether a vector is a certain type with is.___() functions, such as is.character(). is.character(c("hello", "there")) ## [1] TRUE For NA, the test will return a vector testing each element, because NA can be mixed into other values: is.na(c(34, NA)) ## [1] FALSE TRUE We can coerce vectors from one type to the other with as.___() functions, such as as.numeric() as.numeric(c("23", "45")) ## [1] 23 45 as.numeric(c(TRUE, FALSE)) ## [1] 1 0 It is common to have metadata attributes, such as names, attached to R data structures. x = c(1, 2, 3) names(x) = c("a", "b", "c") x ## a b c ## 1 2 3 We can look for more general attributes via the attributes() function: attributes(x) ## $names ## [1] "a" "b" "c" 2.4.1 Ways to subset a vector Positive numeric vector Negative numeric vector performs exclusion Logical vector 2.4.2 Practice implicit subsetting How do you subset the following vector so that it only has positive values? data = c(2, 4, -1, -3, 2, -1, 10) data[data > 0] ## [1] 2 4 2 10 How do you subset the following vector so that it has doesn’t have the character “temp”? chars = c("temp", "object", "temp", "wish", "bumblebee", "temp") chars[chars != "temp"] ## [1] "object" "wish" "bumblebee" How do you subset the following vector so that it has no NA values? vec_with_NA = c(2, 4, NA, NA, 3, NA) vec_with_NA[!is.na(vec_with_NA)] ## [1] 2 4 3 2.5 Factors Factors are a type of vector that holds categorical information, such as sex, gender, or cancer subtype. They are useful for: When you know you have a fixed number of categories. When you want to display character vectors in a non-alphabetical order, which is common in plotting. Inputs for statistical models, as factors are a special type of numerical vectors. place = factor(c("first", "third", "third", "second", "second", "fourth")) place ## [1] first third third second second fourth ## Levels: first fourth second third df = data.frame(p = place) ggplot(df) + geom_bar(aes(x = p)) We can construct ordered factors: place = ordered(c("first", "third", "third", "second","second", "fourth"), levels = c("first", "second", "third", "fourth")) place ## [1] first third third second second fourth ## Levels: first < second < third < fourth df = data.frame(p = place) ggplot(df) + geom_bar(aes(x = p)) 2.6 Dataframes Usually, we load in a dataframe from a spreadsheet or a package, but we can create a new dataframe by using vectors of the same length via the data.frame() function: df = data.frame(x = 1:3, y = c("cup", "mug", "jar")) attributes(df) ## $names ## [1] "x" "y" ## ## $class ## [1] "data.frame" ## ## $row.names ## [1] 1 2 3 library(palmerpenguins) attributes(penguins) ## $class ## [1] "tbl_df" "tbl" "data.frame" ## ## $row.names ## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 ## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 ## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 ## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 ## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 ## [91] 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 ## [109] 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 ## [127] 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 ## [145] 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 ## [163] 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 ## [181] 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 ## [199] 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 ## [217] 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 ## [235] 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 ## [253] 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 ## [271] 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 ## [289] 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 ## [307] 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 ## [325] 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 ## [343] 343 344 ## ## $names ## [1] "species" "island" "bill_length_mm" ## [4] "bill_depth_mm" "flipper_length_mm" "body_mass_g" ## [7] "sex" "year" Why are row names undesirable? Sometimes, data frames will be in a format called “tibble”, as shown in the penguins class names as “tbl_df”, and “tbl”. 2.6.1 Subsetting dataframes Getting one single column: penguins$bill_length_mm ## [1] 39.1 39.5 40.3 NA 36.7 39.3 38.9 39.2 34.1 42.0 37.8 37.8 41.1 38.6 34.6 ## [16] 36.6 38.7 42.5 34.4 46.0 37.8 37.7 35.9 38.2 38.8 35.3 40.6 40.5 37.9 40.5 ## [31] 39.5 37.2 39.5 40.9 36.4 39.2 38.8 42.2 37.6 39.8 36.5 40.8 36.0 44.1 37.0 ## [46] 39.6 41.1 37.5 36.0 42.3 39.6 40.1 35.0 42.0 34.5 41.4 39.0 40.6 36.5 37.6 ## [61] 35.7 41.3 37.6 41.1 36.4 41.6 35.5 41.1 35.9 41.8 33.5 39.7 39.6 45.8 35.5 ## [76] 42.8 40.9 37.2 36.2 42.1 34.6 42.9 36.7 35.1 37.3 41.3 36.3 36.9 38.3 38.9 ## [91] 35.7 41.1 34.0 39.6 36.2 40.8 38.1 40.3 33.1 43.2 35.0 41.0 37.7 37.8 37.9 ## [106] 39.7 38.6 38.2 38.1 43.2 38.1 45.6 39.7 42.2 39.6 42.7 38.6 37.3 35.7 41.1 ## [121] 36.2 37.7 40.2 41.4 35.2 40.6 38.8 41.5 39.0 44.1 38.5 43.1 36.8 37.5 38.1 ## [136] 41.1 35.6 40.2 37.0 39.7 40.2 40.6 32.1 40.7 37.3 39.0 39.2 36.6 36.0 37.8 ## [151] 36.0 41.5 46.1 50.0 48.7 50.0 47.6 46.5 45.4 46.7 43.3 46.8 40.9 49.0 45.5 ## [166] 48.4 45.8 49.3 42.0 49.2 46.2 48.7 50.2 45.1 46.5 46.3 42.9 46.1 44.5 47.8 ## [181] 48.2 50.0 47.3 42.8 45.1 59.6 49.1 48.4 42.6 44.4 44.0 48.7 42.7 49.6 45.3 ## [196] 49.6 50.5 43.6 45.5 50.5 44.9 45.2 46.6 48.5 45.1 50.1 46.5 45.0 43.8 45.5 ## [211] 43.2 50.4 45.3 46.2 45.7 54.3 45.8 49.8 46.2 49.5 43.5 50.7 47.7 46.4 48.2 ## [226] 46.5 46.4 48.6 47.5 51.1 45.2 45.2 49.1 52.5 47.4 50.0 44.9 50.8 43.4 51.3 ## [241] 47.5 52.1 47.5 52.2 45.5 49.5 44.5 50.8 49.4 46.9 48.4 51.1 48.5 55.9 47.2 ## [256] 49.1 47.3 46.8 41.7 53.4 43.3 48.1 50.5 49.8 43.5 51.5 46.2 55.1 44.5 48.8 ## [271] 47.2 NA 46.8 50.4 45.2 49.9 46.5 50.0 51.3 45.4 52.7 45.2 46.1 51.3 46.0 ## [286] 51.3 46.6 51.7 47.0 52.0 45.9 50.5 50.3 58.0 46.4 49.2 42.4 48.5 43.2 50.6 ## [301] 46.7 52.0 50.5 49.5 46.4 52.8 40.9 54.2 42.5 51.0 49.7 47.5 47.6 52.0 46.9 ## [316] 53.5 49.0 46.2 50.9 45.5 50.9 50.8 50.1 49.0 51.5 49.8 48.1 51.4 45.7 50.7 ## [331] 42.5 52.2 45.2 49.3 50.2 45.6 51.9 46.8 45.7 55.8 43.5 49.6 50.8 50.2 I want to select columns bill_length_mm, bill_depth_mm, species, and filter for species that are “Gentoo”: penguins_select = select(penguins, bill_length_mm, bill_depth_mm, species) penguins_gentoo = filter(penguins_select, species == "Gentoo") or penguins_select_2 = penguins[, c("bill_length_mm", "bill_depth_mm", "species")] penguins_gentoo_2 = penguins_select_2[penguins$species == "Gentoo" ,] or penguins_gentoo_2 = penguins_select_2[penguins$species == "Gentoo", c("bill_length_mm", "bill_depth_mm", "species")] I want to filter out rows that have NAs in the column bill_length_mm: penguins_clean = filter(penguins, !is.na(bill_length_mm)) or penguins_clean = penguins[!is.na(penguins$bill_depth_mm) ,] 2.7 Lists Lists operate similarly as vectors as they group data into one dimension, but each element of a list can be any data type or data structure! l1 = list( 1:3, "a", c(TRUE, FALSE, TRUE), c(2.3, 5.9) ) Unlike vectors, you access the elements of a list via the double bracket [[]]. You access a smaller list with single bracket []. (More discussion on the different uses of the bracket here.) Here’s a nice metaphor: If list x is a train carrying objects, then x[[5]] is the object in car 5; x[4:6] is a train of cars 4-6. l1[[1]] ## [1] 1 2 3 l1[[1]][2] ## [1] 2 Use unlist() to coerce a list into a vector. Notice all the automatic coersion that happened for the elements. unlist(l1) ## [1] "1" "2" "3" "a" "TRUE" "FALSE" "TRUE" "2.3" "5.9" We can give names to lists: l1 = list( ranking = 1:3, name = "a", success = c(TRUE, FALSE, TRUE), score = c(2.3, 5.9) ) #or names(l1) = c("ranking", "name", "success", "score") And access named elements of lists via the $ operation: l1$score ## [1] 2.3 5.9 Therefore, l1$score is the same as l1[[4]] and is the same as l1[[\"score\"]]. A dataframe is just a named list of vectors of same length with attributes of (column) names and row.names. 2.8 Matrix A matrix holds information of the same data type in two dimensions - it’s like a two dimensional vector. Matricies are most often used in statistical computing and matrix algebra, such as creating a design matrix. They are often created by taking a vector and reshaping it with a set number of rows and columns, or converting from a dataframe with only one data type. my_matrix = matrix(1:10, nrow = 2) my_matrix ## [,1] [,2] [,3] [,4] [,5] ## [1,] 1 3 5 7 9 ## [2,] 2 4 6 8 10 You access elements of a matrix similar to that of a dataframe’s indexing: #column 3 my_matrix[, 3] ## [1] 5 6 #row 2 my_matrix[2 ,] ## [1] 2 4 6 8 10 #column 3, row 2 my_matrix[2, 3] ## [1] 6 "],["fundamentals-exercises.html", "Chapter 3 Fundamentals Exercises 3.1 Part 1: Vectors 3.2 Part 2: Lists", " Chapter 3 Fundamentals Exercises 3.1 Part 1: Vectors How do you subset the following vector to the first three elements? measurements = c(2, 4, -1, -3, 2, -1, 10) How do you subset the original vector so that it only has negative values? How do you subset the following vector so that it has no NA values? vec_with_NA = c(2, 4, NA, NA, 3, NA) Consider the following logical vector some_logicals. Convert Logical vector -> Numeric vector -> Character vector in two steps. Check that you are doing this correctly along the way by using the class() function, or is.numeric() and is.character(), on the converted data. some_logicals = c(TRUE, TRUE, TRUE, FALSE, TRUE, FALSE) 3.2 Part 2: Lists Consider the following lists with names. patient = list( name = " ", age = 34, pronouns = c("he", "him", "/", "they", "them"), vaccines = c("hep-B", "chickenpox", "HPV"), visits = NA ) visit1 = list( symptoms = c("runny nose", "sore throat", "frustration"), prescription = "recommended time off from work, rest.", date = "1/1/2000" ) visit2 = list( symptoms = c("fainted", "pale complexion"), prescription = "drink water and take time off work.", date = "1/1/2001" ) Access the first element of patient via double brackets [[ ]] and modify it to a value of your choice. Access the named element “pronouns” of patient via double bracket [[ ]] or $ and modify its value so that it doesn’t contain the “/” element. (Use your vector subsetting skills here after you access the appropriate element from the list.) Create a new list all_visits with elements visit1 and visit2. Yes, we’re making lists within lists! Suppose you want to use all_visits to access visit 1’s symptoms. You would continue the double brackets [[ ]] or $ notation: all_visits[[1]] returns a list, so we access the first element of that list via all_visits[[1]][[1]]. #all_visits[[1]][[1]] #or #ll_visits[[1]][["symptoms"]] #or #ll_visits[[1]]$symptoms How would you use all_visits to access visit 2’s prescription? How would you use all_visits to access visit 2’s symptom element “pale complexion”? Remember, once you access a vector, you would go back to the single bracket [ ] to access its elements. Finally, assign all_visits to patient’s visits. 3.2.1 Part 3: Dataframes (Lists) A dataframe is just a named list of vectors of same length with attributes of (column) names and row.names. library(palmerpenguins) head(penguins) ## # A tibble: 6 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## # ℹ 2 more variables: sex <fct>, year <int> Access the body_mass_g column vector of penguins via the double bracket [[ ]], treating penguins like a list, and compute the mean. Remember to use na.rm = TRUE to remove any NA values: mean(x, na.rm = TRUE) Create a new dataframe penguins_clean, which has no NA values in the body_mass_g column. You need to filter out rows that have NAs in the column bill_length_mm: Now, subset penguins_clean to each of the three species and compute their respective mean value of body_mass_g. Because you already got rid of NAs in body_mass_g, you can just use mean(x) without the extra option. How do they compare? Finally, make a box plot of species (x-axis) vs. body_mass_g (y-axis) via penguins_clean dataframe. I’ll get you started… #ggplot(penguins_clean) + aes(x = , y = ) + geom_boxplot() "],["data-cleaning-part-1.html", "Chapter 4 Data Cleaning, Part 1 4.1 Interpreting functions, carefully 4.2 Recoding Data / Conditionals 4.3 Conditionals", " Chapter 4 Data Cleaning, Part 1 4.1 Interpreting functions, carefully As you become more independent R programmers, you will spend time learning about new functions on your own. We have gone over the basic anatomy of a function call back in Intro to R, but now let’s go a bit deeper to understand how a function is built and how to call them. Recall that a function has a function name, input arguments, and a return value. Function definition consists of assigning a function name with a “function” statement that has a comma-separated list of named function arguments, and a return expression. The function name is stored as a variable in the global environment. In order to use the function, one defines or import it, then one calls it. Example: addFunction = function(num1, num2) { result = num1 + num2 return(result) } result = addFunction(3, 4) When the function is called in line 5, the variables for the arguments are reassigned to function arguments to be used within the function and helps with the modular form. What do you think are some valid inputs for this function? To see why we need the variables of the arguments to be reassigned, consider the following function that is not modular: x = 3 y = 4 addFunction = function(num1, num2) { result = x + y return(result) } result = addFunction(10, -10) Some syntax equivalents on calling the function: addFunction(3, 4) addFunction(num1 = 3, num2 = 4) addFunction(num2 = 4, num1 = 3) but this could be different: addFunction(4, 3) With a deeper knowledge of how functions are built, when you encounter a foreign function, you can look up its help page to understand how to use it. For example, let’s look at mean(): ?mean Arithmetic Mean Description: Generic function for the (trimmed) arithmetic mean. Usage: mean(x, ...) ## Default S3 method: mean(x, trim = 0, na.rm = FALSE, ...) Arguments: x: An R object. Currently there are methods for numeric/logical vectors and date, date-time and time interval objects. Complex vectors are allowed for ‘trim = 0’, only. trim: the fraction (0 to 0.5) of observations to be trimmed from each end of ‘x’ before the mean is computed. Values of trim outside that range are taken as the nearest endpoint. na.rm: a logical evaluating to ‘TRUE’ or ‘FALSE’ indicating whether ‘NA’ values should be stripped before the computation proceeds. ...: further arguments passed to or from other methods. Notice that the arguments trim = 0, na.rm = FALSE have default values. This means that these arguments are optional - you should provide it only if you want to. With this understanding, you can use mean() in a new way: numbers = c(1, 2, NA, 4) mean(x = numbers, na.rm = TRUE) ## [1] 2.333333 The use of . . . (dot-dot-dot): This is a special argument that allows a function to take any number of arguments. This isn’t very useful for the mean() function, but it makes sense for function such as select() and filter(), and mutate(). For instance, in select(), once you provide your dataframe for the argument .data, you can pile on as many columns to select in the rest of the argument. Usage: select(.data, ...) Arguments: .data: A data frame, data frame extension (e.g. a tibble), or a lazy data frame (e.g. from dbplyr or dtplyr). See _Methods_, below, for more details. ...: <‘tidy-select’> One or more unquoted expressions separated by commas. Variable names can be used as if they were positions in the data frame, so expressions like ‘x:y’ can be used to select a range of variables. You will look at the function documentation on your own to see how to deal with more complex cases. 4.2 Recoding Data / Conditionals It is often said that 80% of data analysis is spent on the cleaning and preparing data. Today we will start looking at common data cleaning tasks. Suppose that you have a column in your data that needs to be recoded. Since a dataframe’s column, when selected via $, is a vector, let’s start talking about recoding vectors. If we have a numeric vector, then maybe you want to have certain values to be out of bounds, or assign a range of values to a character category. If we have a character vector, then maybe you want to reassign it to a different value. Here are popular recoding logical scenarios: If: “If elements of the vector meets condition, then they are assigned value.” If-else: “If elements of the vector meets condition, then they are assigned value X. Otherwise, they are assigned value Y.” If-else_if-else: “If elements of the vector meets condition A, then they are assigned value X. Else, if the elements of the vector meets condition B, they are assigned value Y. Otherwise, they are assigned value Z.” Let’s look at a vector of grade values, as an example: grade = c(90, 78, 95, 74, 56, 81, 102) If Instead of having the bracket [ ] notation on the right hand side of the equation, if it is on the left hand side of the equation, then we can modify a subset of the vector. grade1 = grade grade1[grade1 > 100] = 100 If-else grade2 = if_else(grade > 60, TRUE, FALSE) If-else_if-else grade3 = case_when(grade >= 90 ~ "A", grade >= 80 ~ "B", grade >= 70 ~ "C", grade >= 60 ~ "D", .default = "F") Let’s do it for dataframes now. simple_df = data.frame(grade = c(90, 78, 95, 74, 56, 81, 102), status = c("case", " ", "Control", "control", "Control", "Case", "case")) If simple_df1 = simple_df simple_df1$grade[simple_df1$grade > 100] = 100 If-else simple_df2 = simple_df simple_df2$grade = ifelse(simple_df2$grade > 60, TRUE, FALSE) or simple_df2 = mutate(simple_df, grade = ifelse(grade > 60, TRUE, FALSE)) If-else_if-else simple_df3 = simple_df simple_df3$grade = case_when(simple_df3$grade >= 90 ~ "A", simple_df3$grade >= 80 ~ "B", simple_df3$grade >= 70 ~ "C", simple_df3$grade >= 60 ~ "D", .default = "F") or simple_df3 = mutate(simple_df, grade = case_when(grade >= 90 ~ "A", grade >= 80 ~ "B", grade >= 70 ~ "C", grade >= 60 ~ "D", .default = "F")) 4.3 Conditionals The 3 common scenarios we looked at for recoding data is closely tied to the concept of conditionals in programming: given certain conditions, you run a specific code chunk. Given a vector’s value, assign it a different value. Or, given a value, run the following hundred lines of code. Here is what it looks like: If: if(expression_is_TRUE) { #code goes here } If-else: if(expression_is_TRUE) { #code goes here }else { #other code goes here } If-else_if-else: if(expression_A_is_TRUE) #code goes here }else if(expression_B_is_TRUE) { #other code goes here }else { #some other code goes here } The expression that is being tested whether it is TRUE must be a singular logical value, and not a logical vector. If the latter, see the recoding section for now. Example: nuc = "A" if(nuc == "A") { nuc = "T" }else if(nuc == "T") { nuc = "A" }else if(nuc == "C") { nuc = "C" }else if(nuc == "G") { nuc = "C" }else { nuc = NA } nuc ## [1] "T" Example: my_input = c(1, 3, 5, 7, 9) #my_input = c("e", "e", "a", "i", "o") if(is.numeric(my_input)) { result = mean(my_input) }else if(is.character(my_input)) { result = table(my_input) } result ## [1] 5 "],["data-cleaning-part-1-exercises.html", "Chapter 5 Data Cleaning, Part 1 Exercises 5.1 Part 1: Looking at documentation to load in data 5.2 Part 2: Recoding data: warm-up 5.3 Part 3: Recoding data in State Cancer Profiles", " Chapter 5 Data Cleaning, Part 1 Exercises 5.1 Part 1: Looking at documentation to load in data Suppose that you want to load in data “students.csv” in a CSV format, and you don’t know what tools to use. You decide to see whether the package “readr” can be useful to solve your problem. Where should you look? All R packages must be stored on CRAN (Comprehensive R Archive Network), and all packages have a website that points to the reference manual (what is pulled up using the ? command), source code, vignettes examples, and dependencies on other packages. Here is the website for “readr”. In the package, you find some potential functions for importing your data: read_csv(\"file.csv\") for comma-separated files read_tsv(\"file.tsv\") for tab-deliminated files read_excel(\"example.xlsx\") for excel files read_excel(\"example.xlsx\", sheet = \"sheet1\") for excel files with a sheet name read_delim() for general-deliminated files, such as: read_delim(\"file.csv\", sep = \",\"). After looking at the vignettes, it seems that read_csv() is a function to try. Let’s look at the read_csv() function documentation, which can be accessed via ?read_csv. read_csv( file, col_names = TRUE, col_types = NULL, col_select = NULL, id = NULL, locale = default_locale(), na = c("", "NA"), quoted_na = TRUE, quote = "\\"", comment = "", trim_ws = TRUE, skip = 0, n_max = Inf, guess_max = min(1000, n_max), name_repair = "unique", num_threads = readr_threads(), progress = show_progress(), show_col_types = should_show_types(), skip_empty_rows = TRUE, lazy = should_read_lazy() ) We see that the only required argument is the file variable, which is documented to be “Either a path to a file, a connection, or literal data (either a single string or a raw vector).” All the other arguments are considered optional, because they have a pre-allocated value in the documentation. Load in “students.csv” via read_csv() function as a dataframe variable students and take a look at its contents via View(). library(tidyverse) ## Warning: package 'tidyverse' was built under R version 4.0.3 ## ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ── ## ✔ ggplot2 3.3.2 ✔ purrr 0.3.4 ## ✔ tibble 3.2.1 ✔ dplyr 1.0.2 ## ✔ tidyr 1.1.2 ✔ stringr 1.4.0 ## ✔ readr 1.4.0 ✔ forcats 0.5.0 ## Warning: package 'purrr' was built under R version 4.0.5 ## Warning: package 'stringr' was built under R version 4.0.3 ## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ── ## ✖ dplyr::filter() masks stats::filter() ## ✖ dplyr::lag() masks stats::lag() Something looks weird here. There is only one column, and it seems that the first two entries start with “#”, and don’t fit a CSV file format. These first two entries that start with “#” likely are comments giving metadata about the file, and they should be ignore when loading in the data. Let’s try again. Take a look at the documentation for the comment argument and give it a character value \"#\" with this situation. Any text after the comment characters will be silently ignored. The column names are not very consistent . Take a look at the documentation for the col_names argument and give it a value of c(\"student_id\", \"full_name\", \"favorite_food\", \"meal_plan\", \"age\"). Alternatively, you could have loaded the data in without col_names option and modified the column names by accessing names(students). For more information on loading in data, see this chapter of R for Data Science. 5.2 Part 2: Recoding data: warm-up Consider this vector: scores = c(23, 46, -3, 5, -1) Recode scores so that all the negative values are 0. Let’s look at the values of students dataframe more carefully. We will do some recoding on this small dataframe. It may feel trivial because you could do this by hand in Excel, but this is a practice on how we can scale this up with larger datasets! Notice that some of the elements of this dataframe has proper NA values and also a character “N/A”. We want “N/A” to be a proper NA value. Recode “N/A” to NA in the favorite_food column: Recode “five” to 5 in the age column: Create a new column age_category so that it has value “toddler” if age is < 6, and “child” if age is >= 6. (Hint: You can create a new column via mutate, or you can directly refer to the new column via student$``age_category.) Create a new column favorite_food_numeric so that it has value 1 if favorite_food is “Breakfast and lunch”, 2 if “Lunch only”, and 3 if “Dinner only”. 5.3 Part 3: Recoding data in State Cancer Profiles Starting from this exercise, we will start building out an end-to-end analysis using data from the National Cancer Institute’s State Cancer Profile: [State Cancer Profile data] was developed with the idea to provide a geographic profile of cancer burden in the United States and reveal geographic disparities in cancer incidence, mortality, risk factors for cancer, and cancer screening, across different population subgroups. In this analysis, we want to examine cancer incidence rates in state of Washington and make some comparisons between groups. The cancer incidence rate can be accessed at this website, once you give input of what kind of incidence data you want to access. If you want to analyze this data in R, it takes a bit of work of exporting the data and loading it into R. To access this data easier in R, DaSL staff built a R package cancerprof to easily load in the data. Let’s look at the package’s documentation to see how to get access to cancer incidence data. Towards the bottom of the documentation are some useful examples to consider as starting point. Load in data about the following population: melanoma incidence in WA at the county level for males of all ages, all cancer stages, averaged in the past 5 years. Store it as a dataframe variable named melanoma_incidence (If you are stuck, you can use the first example in the documentation.) Take a look at the data yourself and explore it. Let’s select a few columns of interest and give them column names that doesn’t contain spaces. We can access column names with spaces via the backtick ` symbol. #uncomment to run! #melanoma_incidence = select(melanoma_incidence, County, `Age Adjusted Incidence Rate`, `Recent Trend`) #names(melanoma_incidence) = c("County", "Age_adjusted_incidence_rate", "Recent_trend") Take a look at the column Age_adjusted_incidence_rate. It has missing data coded as “*” (notice the space after *). Recode “*” as NA. Finally, notice that the data type for Age_adjusted_incidence_rate is character, if you run the function is.character() or class() on it. Convert it to a numeric data type. "],["data-cleaning-part-2.html", "Chapter 6 Data Cleaning, Part 2 6.1 Tidy Data 6.2 Uses of Tidy data 6.3 Subjectivity in Tidy Data 6.4 References", " Chapter 6 Data Cleaning, Part 2 library(tidyverse) 6.1 Tidy Data It is important to have standard of organizing data, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of Tidy data, developed by Hadley Wickham: Each variable must have its own column. Each observation must have its own row. Each value must have its own cell. If we want to be technical about what variables and observations are, Hadley Wickham describes: A dataset is a collection of values, usually either numbers (if quantitative) or strings (if qualitative). Every value belongs to a variable and an observation. A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units. An observation contains all values measured on the same unit (like a person, or a day, or a race) across attributes. A tidy dataframe. Besides a standard, Tidy data is useful because many tools in R are most effective when your data is in a Tidy format. This includes data visualization with ggplot, regression models, databases, and more. These tools assumes the values of each variable fall in their own column vector. It seems hard to go wrong with these simple criteria of Tidy data! However, in reality, many dataframes we load in aren’t Tidy, and it’s easiest seen through counterexamples and how to fix it. Here are some common ways that data becomes un-Tidy: Columns contain values of variables, rather than variables Variables are stored in rows Multiple variables are stored in a single column After some clear examples, we emphasize that “Tidy” data is subjective to what kind of analysis you want to do with the dataframe. 6.1.1 1. Columns contain values, rather than variables (Long is tidy) df = data.frame(Store = c("A", "B"), Year = c(2018, 2018), Q1_Sales = c(55, 98), Q2_Sales = c(45, 70), Q3_Sales = c(22, 60), Q4_Sales = c(50, 60)) df ## Store Year Q1_Sales Q2_Sales Q3_Sales Q4_Sales ## 1 A 2018 55 45 22 50 ## 2 B 2018 98 70 60 60 Each observation is a store, and each observation has its own row. That looks good. The columns “Q1_Sales”, …, “Q4_Sales” seem to be values of a single variable “quarter” of our observation. The values of “quarter” are not in a single column, but are instead in the columns. df_long = pivot_longer(df, c("Q1_Sales", "Q2_Sales", "Q3_Sales", "Q4_Sales"), names_to = "quarter", values_to = "sales") df_long ## # A tibble: 8 × 4 ## Store Year quarter sales ## <chr> <dbl> <chr> <dbl> ## 1 A 2018 Q1_Sales 55 ## 2 A 2018 Q2_Sales 45 ## 3 A 2018 Q3_Sales 22 ## 4 A 2018 Q4_Sales 50 ## 5 B 2018 Q1_Sales 98 ## 6 B 2018 Q2_Sales 70 ## 7 B 2018 Q3_Sales 60 ## 8 B 2018 Q4_Sales 60 Now, each observation is a store’s quarter, and each observation has its own row. The new columns “quarter” and “sales” are variables that describes our observation, and describes our values. We’re in a tidy state! We have transformed our data to a “longer” format, as our observation represents something more granular or detailed than before. Often, the original variables values will repeat itself in a “longer format”. We call the previous state of our dataframe is a “wider” format. 6.1.2 2. Variables are stored in rows (Wide is tidy) Are all tidy dataframes Tidy in a “longer” format? df2 = data.frame(Sample = c("A", "B"), KRAS_mutation = c(TRUE, FALSE), KRAS_expression = c(2.3, 3.9)) df2 ## Sample KRAS_mutation KRAS_expression ## 1 A TRUE 2.3 ## 2 B FALSE 3.9 Each observation is a sample, and each observation has its own row. Looks good. Each variable has its own column, and no values are in columns. What happens if we make it longer? df2_long = pivot_longer(df2, c("KRAS_mutation", "KRAS_expression"), names_to = "gene", values_to = "values") df2_long ## # A tibble: 4 × 3 ## Sample gene values ## <chr> <chr> <dbl> ## 1 A KRAS_mutation 1 ## 2 A KRAS_expression 2.3 ## 3 B KRAS_mutation 0 ## 4 B KRAS_expression 3.9 Here, each observation is a sample’s gene…type? The observation feels awkward because variables are stored in rows. Also, the column “values” contains multiple variable types: gene expression and mutation values that got coerced to numeric! To make this dataframe wider, df2_long_wide = pivot_wider(df2_long, names_from = "gene", values_from = "values") df2_long_wide$KRAS_mutation = as.logical(df2_long_wide$KRAS_mutation) df2_long_wide ## # A tibble: 2 × 3 ## Sample KRAS_mutation KRAS_expression ## <chr> <lgl> <dbl> ## 1 A TRUE 2.3 ## 2 B FALSE 3.9 We are back to our orignal form, and it was already Tidy. 6.1.3 3. Multiple variables are stored in a single column table3 ## # A tibble: 6 × 3 ## country year rate ## * <chr> <int> <chr> ## 1 Afghanistan 1999 745/19987071 ## 2 Afghanistan 2000 2666/20595360 ## 3 Brazil 1999 37737/172006362 ## 4 Brazil 2000 80488/174504898 ## 5 China 1999 212258/1272915272 ## 6 China 2000 213766/1280428583 There seems to be two variables in the numerator and denominator of “rate” column. Let’s separate it. separate(table3, col = "rate", into = c("count", "population"), sep = "/") ## # A tibble: 6 × 4 ## country year count population ## <chr> <int> <chr> <chr> ## 1 Afghanistan 1999 745 19987071 ## 2 Afghanistan 2000 2666 20595360 ## 3 Brazil 1999 37737 172006362 ## 4 Brazil 2000 80488 174504898 ## 5 China 1999 212258 1272915272 ## 6 China 2000 213766 1280428583 6.2 Uses of Tidy data In general, many functions for analysis and visualization in R assumes that the input dataframe is Tidy. These tools assumes the values of each variable fall in their own column vector. For instance, from our first example, we can compare sales across quarters and stores. df_long ## # A tibble: 8 × 4 ## Store Year quarter sales ## <chr> <dbl> <chr> <dbl> ## 1 A 2018 Q1_Sales 55 ## 2 A 2018 Q2_Sales 45 ## 3 A 2018 Q3_Sales 22 ## 4 A 2018 Q4_Sales 50 ## 5 B 2018 Q1_Sales 98 ## 6 B 2018 Q2_Sales 70 ## 7 B 2018 Q3_Sales 60 ## 8 B 2018 Q4_Sales 60 ggplot(df_long) + aes(x = quarter, y = sales, group = Store) + geom_point() + geom_line() Although in its original state we can also look at sales between quarter, we can only look between two quarters at once. Tidy data encourages looking at data in the most granular scale. ggplot(df) + aes(x = Q1_Sales, y = Q2_Sales, color = Store) + geom_point() 6.3 Subjectivity in Tidy Data We have looked at clear cases of when a dataset is Tidy. In reality, the Tidy state depends on what we call variables and observations. kidney = data.frame(stone_size = c("Small", "Large"), treatment.A_recovered = c(81, 192), treatment.A_failed = c(6, 71), treatment.B_recovered = c(234, 55), treatment.B_failed = c(36, 25)) kidney ## stone_size treatment.A_recovered treatment.A_failed treatment.B_recovered ## 1 Small 81 6 234 ## 2 Large 192 71 55 ## treatment.B_failed ## 1 36 ## 2 25 Right now, the kidney dataframe clearly has values of a variable in the column. Let’s try to make it Tidy by making it into a longer form and separating out variables that are together in a column. kidney_long = pivot_longer(kidney, c("treatment.A_recovered", "treatment.A_failed", "treatment.B_recovered", "treatment.B_failed"), names_to = "treatment_outcome", values_to = "count") kidney_long = separate(kidney_long, "treatment_outcome", c("treatment", "outcome"), "_") kidney_long ## # A tibble: 8 × 4 ## stone_size treatment outcome count ## <chr> <chr> <chr> <dbl> ## 1 Small treatment.A recovered 81 ## 2 Small treatment.A failed 6 ## 3 Small treatment.B recovered 234 ## 4 Small treatment.B failed 36 ## 5 Large treatment.A recovered 192 ## 6 Large treatment.A failed 71 ## 7 Large treatment.B recovered 55 ## 8 Large treatment.B failed 25 Here, each observation is a kidney stone’s treatment’s outcome type, and each observation has its own row. The column “count” describes our observation, and describes our values. This dataframe seems reasonably Tidy. How about this? kidney_long_still = pivot_wider(kidney_long, names_from = "outcome", values_from = "count") kidney_long_still ## # A tibble: 4 × 4 ## stone_size treatment recovered failed ## <chr> <chr> <dbl> <dbl> ## 1 Small treatment.A 81 6 ## 2 Small treatment.B 234 36 ## 3 Large treatment.A 192 71 ## 4 Large treatment.B 55 25 Here, each observation is a kidney stone’s treatment, and each observation has its own row. The columns “recovered” and “failed” are variables that describes our observation, and describes its corresponding values. This dataframe seems reasonably Tidy, also. The reason why both of these versions seem Tidy is that the columns “recovered” and “failed” can be interpreted as independent variables and values of the variable “treatment”. Ultimately, we decide which dataframe we prefer based on the analysis we want to do. For instance, when our observation is about a kidney stone’s treatment’s outcome type, we compare it between outcome type, treatment, and stone size. ggplot(kidney_long) + aes(x = treatment, y = count, fill = outcome) + geom_bar(position="dodge", stat="identity") + facet_wrap(~stone_size) When our observation is about a kidney stone’s treatment’s, we compare a new variable recovery rate ( = recovered / (recovered + failed)) between treatment and stone size. kidney_long_still = mutate(kidney_long_still, recovery_rate = recovered / (recovered + failed)) ggplot(kidney_long_still) + aes(x = treatment, y = recovery_rate, fill = stone_size) + geom_bar(position="dodge", stat="identity") 6.4 References https://vita.had.co.nz/papers/tidy-data.html https://kiwidamien.github.io/what-is-tidy-data.html "],["data-cleaning-part-2-exercises.html", "Chapter 7 Data Cleaning, Part 2 Exercises", " Chapter 7 Data Cleaning, Part 2 Exercises "],["writing-your-first-function.html", "Chapter 8 Writing your first function", " Chapter 8 Writing your first function Function machine from algebra class. We write functions for two main, often overlapping, reasons: Following DRY (Don’t Repeat Yourself) principle: If you find yourself repeating similar patterns of code, you should write a function that executes that pattern. This saves time and the risk of mistakes. Create modular structure and abstraction: Having all of your code in one place becomes increasingly complicated as your program grows. Think of the function as a mini-program that can perform without the rest of the program. Organizing your code by functions gives modular structure, as well as abstraction: you only need to know the function name, inputs, and output to use it and don’t have to worry how it works. Some advice on writing functions: Code that has a well-defined set of inputs and outputs make a good function. A function should do only one, well-defined task. 8.0.1 Anatomy of a function definition Function definition consists of assigning a function name with a “function” statement that has a comma-separated list of named function arguments, and a return expression. The function name is stored as a variable in the global environment. In order to use the function, one defines or import it, then one calls it. Example: addFunction = function(argument1, argument2) { result = argument1 + argument2 return(result) } z = addFunction(3, 4) With function definitions, not all code runs from top to bottom. The first four lines defines the function, but the function is never run. It is called on line 5, and the lines within the function are executed. When the function is called in line 5, the variables for the arguments are reassigned to function arguments to be used within the function and helps with the modular form. We need to introduce the concept of local and global environments to distinguish variables used only for a function from variables used for the entire program. 8.0.2 Local and global environments { } represents variable scoping: within each { }, if variables are defined, they are stored in a local environment, and is only accessible within { }. All function arguments are stored in the local environment. The overall environment of the program is called the global environment and can be also accessed within { }. The reason of having some of this “privacy” in the local environment is to make functions modular - they are independent little tools that should not interact with the rest of the global environment. Imagine someone writing a tool that they want to give someone else to use, but the tool depends on your environment, vice versa. 8.0.3 A step-by-step example Using the addFunction function, let’s see step-by-step how the R interpreter understands our code: We define the function in the global environment. We call the function, and the function arguments 3, 4 are assigned to argument1 and argument2, respectively in the function’s local environment. We run the first line of code in the function body. The new variable “result” is stored in the local environment because it is within { }. We run the second line of code in the function body to return a value. The return value from the function is assigned to the variable z in the global environment. All local variables for the function are erased now that the function call is over. 8.0.4 Function arguments create modularity First time writers of functions might ask: why are variables we use for the arguments of a function reassigned for function arguments in the local environment? Here is an example when that process is skipped - what are the consequences? x = 3 y = 4 addFunction = function(argument1, argument2) { result = x + y return(result) } z = addFunction(x, y) w = addFunction(10, -5) What do you expect the value of z to be? How about w? Here is the execution for w: We define the variables and function in the global environment. We call the function, and the function arguments 10, -5 are assigned to argument1 and argument2, respectively in the function’s local environment. We run the first line of code in the function body. The new variable “result” is stored in the local environment because it is within { }. We run the second line of code in the function body to return a value. The return value from the function is assigned to the variable w in the global environment. All local variables for the function are erased now that the function call is over. The function did not work as expected because we used hard-coded variables from the global environment and not function argument variables unique to the function use! 8.0.5 Exercises Create a function, called add_and_raise_power in which the function takes in 3 numeric arguments. The function computes the following: the first two arguments are added together and raised to a power determined by the 3rd argument. The function returns the resulting value. Here is a use case: add_and_raise_power(1, 2, 3) = 27 because the function will return this expression: (1 + 2) ^ 3. Another use case: add_and_raise_power(3, 1, 2) = 16 because of the expression (3 + 1) ^ 2. Confirm with that these use cases work. Can this function used for numeric vectors? add_and_raise_power = function(x, y, z) { result = (x + y)^z return(result) } add_and_raise_power(1, 2, 3) ## [1] 27 Create a function, called my_dim in which the function takes in one argument: a dataframe. The function returns the following: a length-2 numeric vector in which the first element is the number of rows in the dataframe, and the second element is the number of columns in the dataframe. Your result should be identical as the dim function. How can you leverage existing functions such as nrow and ncol? Use case: my_dim(penguins) = c(344, 8) library(palmerpenguins) my_dim = function(df) { result = c(nrow(df), ncol(df)) return(result) } my_dim(penguins) ## [1] 344 8 Create a function, called medicaid_eligible in which the function takes in one argument: a numeric vector called age. The function returns a numeric vector with the same length as age, in which elements are 0 for indicies that are less than 65 in age, and 1 for indicies 65 or higher in age. Use cases: medicaid_eligible(c(30, 70)) = c(0, 1) medicaid_eligible = function(age) { result = age result[age < 65] = 0 result[age >= 65] = 1 return(result) } medicaid_eligible(c(30, 70)) ## [1] 0 1 "],["functions-exercises.html", "Chapter 9 Functions Exercises", " Chapter 9 Functions Exercises "],["repeating-tasks.html", "Chapter 10 Repeating tasks 10.1 For loops 10.2 Functionals 10.3 Case studies", " Chapter 10 Repeating tasks Suppose that you want to repeat a chunk of code many times, but changing one variable’s value each time you do it. This could be modifying each element of a vector with the same operation, or analyzing a dataframe with different parameters. There are three common strategies to go about this: Copy and paste the code chunk, and change that variable’s value. Repeat. This can be a starting point in your analysis, but will lead to errors easily. Use a for loop to repeat the chunk of code, and let it loop over the changing variable’s value. This is popular for many programming languages, but the R programming culture encourages a functional way instead. Functionals allow you to take a function that solves the problem for a single input and generalize it to handle any number of inputs. This is very popular in R programming culture. 10.1 For loops A for loop repeats a chunk of code many times, once for each element of an input vector. for (my_element in my_vector) { chunk of code } Most often, the “chunk of code” will make use of my_element. 10.1.0.1 We can loop through elements of a vector and print it out. my_vector = c(1, 3, 5, 7) for(my_element in my_vector) { print(my_element) } ## [1] 1 ## [1] 3 ## [1] 5 ## [1] 7 10.1.0.2 Alternatively, we can loop through the indicies of a vector and print it out. The function seq_along() creates the indicies of a vector. for(i in seq_along(my_vector)) { print(my_vector[i]) } ## [1] 1 ## [1] 3 ## [1] 5 ## [1] 7 10.2 Functionals A functional is a function that takes in a data structure and function as inputs and applies the function on the data structure, element by element. It maps your input data structure to an output data structure based on the function. It encourages the usage of modular functions in your code. Or, We will use the purrr package in tidyverse to use functionals. map() takes in a vector or a list, and then applies the function on each element of it. The output is always a list. (You see the output twice, because it prints out element by element, and then returns a list.) map(my_vector, print) ## [1] 1 ## [1] 3 ## [1] 5 ## [1] 7 ## [[1]] ## [1] 1 ## ## [[2]] ## [1] 3 ## ## [[3]] ## [1] 5 ## ## [[4]] ## [1] 7 Lists are useful if what you are using it on requires a flexible data structure. To be more specific about the output type, you can do this via the map_* function, where * specifies the output type: map_lgl(), map_chr(), and map_dbl() functions return vectors of logical values, strings, or numbers respectively. For example, to make sure your output is a double (numeric): map_dbl(my_vector, log) ## [1] 0.000000 1.098612 1.609438 1.945910 All of these are toy examples that gets us familiar with the syntax, but we already have built-in functions to solve these problems. Let’s see some real-life case studies. 10.3 Case studies 10.3.1 1. Loading in multiple dataframes from files for analysis Suppose that we want to load in a few dataframes, and store them in a list of dataframes for analysis downstream. We start with the filepaths we want to load in as dataframes. paths = c("classroom_data/students.csv", "classroom_data/CCLE_metadata.csv") The function we want to use to load the data in will be read_csv(). Let’s practice writing out one iteration: result = read_csv(paths[1]) 10.3.1.1 To do this functionally, we think about: What variable we need to loop through: paths The repeated task as a function: read_csv() The looping mechanism, and its output: map() outputs lists. loaded_dfs = map(paths, read_csv) 10.3.1.2 To do this with a for loop, we think about: What variable we need to loop through: paths. Do we need to store the outcome of this loop in a data structure? Yes, a list. At each iteration, what are we doing? Use read_csv() on the current element, and store it in the output list. paths = c("classroom_data/students.csv", "classroom_data/CCLE_metadata.csv") loaded_dfs = vector(mode = "list", length = length(paths)) for(i in seq_along(paths)) { df = read_csv(paths[i]) loaded_dfs[[i]] = df } 10.3.2 2. Analyze a dataframe differently with different parameters. Suppose you are working with the penguins dataframe: library(palmerpenguins) head(penguins) ## # A tibble: 6 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## # ℹ 2 more variables: sex <fct>, year <int> and you want to look at the mean bill_length_mm for each of the three species (Adelie, Chinstrap, Gentoo). Let’s practice writing out one iteration: species_to_analyze = c("Adelie", "Chinstrap", "Gentoo") penguins_subset = filter(penguins, species == species_to_analyze[1]) mean(penguins_subset$bill_length_mm, na.rm = TRUE) ## [1] 38.79139 10.3.2.1 To do this functionally, we think about: What variable we need to loop through: c(\"Adelie\", \"Chinstrap\", \"Gentoo\") The repeated task as a function: a custom function that takes in a specie of interest. The function filters the rows of penguins to the species of interest, and compute the mean of bill_length_mm. The looping mechanism, and its output: map_dbl() outputs (double) numeric vectors. analysis = function(current_species) { penguins_subset = dplyr::filter(penguins, species == current_species) return(mean(penguins_subset$bill_length_mm, na.rm=TRUE)) } map_dbl(c("Adelie", "Chinstrap", "Gentoo"), analysis) ## [1] 38.79139 48.83382 47.50488 10.3.2.2 To do this with a for loop, we think about: What variable we need to loop through: c(\"Adelie\", \"Chinstrap\", \"Gentoo\"). Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector. At each iteration, what are we doing? Filter the rows of penguins to the species of interest, and compute the mean of bill_length_mm. outcome = rep(NA, length(species_to_analyze)) for(i in seq_along(species_to_analyze)) { penguins_subset = filter(penguins, species == species_to_analyze[i]) outcome[i] = mean(penguins_subset$bill_length_mm, na.rm=TRUE) } outcome ## [1] 38.79139 48.83382 47.50488 10.3.3 3. Calculate summary statistics on columns of a dataframe. Suppose that you are interested in the numeric columns of the penguins dataframe. penguins_numeric = penguins %>% select(bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g) and you are interested to look at the mean of each column. It is very helpful to interpret the dataframe penguins_numeric as a list, iterating through each column as an element of a list. Let’s practice writing out one iteration: mean(penguins_numeric[[1]], na.rm = TRUE) ## [1] 43.92193 10.3.3.1 To do this functionally, we think about: What variable we need to loop through: the list penguins_numeric The repeated task as a function: mean() with the argument na.rm = TRUE. The looping mechanism, and its output: map_dbl() outputs (double) numeric vectors. map_dbl(penguins_numeric, mean, na.rm = TRUE) ## bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## 43.92193 17.15117 200.91520 4201.75439 Here, R is interpreting the dataframe penguins_numeric as a list, iterating through each column as an element of a list: 10.3.3.2 To do this with a for loop, we think about: What variable we need to loop through: the elements of penguins_numeric as a list. Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector. At each iteration, what are we doing? Compute the mean of an element of penguins_numeric. result = rep(NA, ncol(penguins_numeric)) for(i in seq_along(penguins_numeric)) { result[i] = mean(penguins_numeric[[i]], na.rm = TRUE) } result ## [1] 43.92193 17.15117 200.91520 4201.75439 "],["repetition-exercises.html", "Chapter 11 Repetition Exercises", " Chapter 11 Repetition Exercises "],["about-the-authors.html", "About the Authors", " About the Authors These credits are based on our course contributors table guidelines. Credits Names Pedagogy Lead Content Instructor(s) Chris Lo Lecturer Chris Lo Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved If any other authors besides lead instructor Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved Wrote less than a chapter Content Editor(s)/Reviewer(s) Checked your content Content Director(s) Helped guide the content direction Content Consultants (include chapter name/link in parentheses or word “General”) - make new line if more than one chapter involved Gave high level advice on content Acknowledgments Gave small assistance to content but not to the level of consulting Production Content Publisher(s) Helped with publishing platform Content Publishing Reviewer(s) Reviewed overall content and aesthetics on publishing platform Technical Course Publishing Engineer(s) Helped with the code for the technical aspects related to the specific course generation Template Publishing Engineers Candace Savonen, Carrie Wright, Ava Hoffman Publishing Maintenance Engineer Candace Savonen Technical Publishing Stylists Carrie Wright, Ava Hoffman, Candace Savonen Package Developers (ottrpal) Candace Savonen, John Muschelli, Carrie Wright Art and Design Illustrator(s) Created graphics for the course Figure Artist(s) Created figures/plots for course Videographer(s) Filmed videos Videography Editor(s) Edited film Audiographer(s) Recorded audio Audiography Editor(s) Edited audio recordings Funding Funder(s) Institution/individual who funded course including grant number Funding Staff Staff members who help with funding ## ─ Session info ─────────────────────────────────────────────────────────────── ## setting value ## version R version 4.0.2 (2020-06-22) ## os Ubuntu 20.04.5 LTS ## system x86_64, linux-gnu ## ui X11 ## language (EN) ## collate en_US.UTF-8 ## ctype en_US.UTF-8 ## tz Etc/UTC ## date 2024-03-28 ## ## ─ Packages ─────────────────────────────────────────────────────────────────── ## package * version date lib source ## askpass 1.1 2019-01-13 [1] RSPM (R 4.0.3) ## assertthat 0.2.1 2019-03-21 [1] RSPM (R 4.0.5) ## bookdown 0.24 2024-03-13 [1] Github (rstudio/bookdown@88bc4ea) ## bslib 0.6.1 2023-11-28 [1] CRAN (R 4.0.2) ## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.0.2) ## callr 3.5.0 2020-10-08 [1] RSPM (R 4.0.2) ## cli 3.6.2 2023-12-11 [1] CRAN (R 4.0.2) ## crayon 1.3.4 2017-09-16 [1] RSPM (R 4.0.0) ## desc 1.2.0 2018-05-01 [1] RSPM (R 4.0.3) ## devtools 2.3.2 2020-09-18 [1] RSPM (R 4.0.3) ## digest 0.6.25 2020-02-23 [1] RSPM (R 4.0.0) ## ellipsis 0.3.1 2020-05-15 [1] RSPM (R 4.0.3) ## evaluate 0.23 2023-11-01 [1] CRAN (R 4.0.2) ## fansi 0.4.1 2020-01-08 [1] RSPM (R 4.0.0) ## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.0.2) ## fs 1.5.0 2020-07-31 [1] RSPM (R 4.0.3) ## glue 1.4.2 2020-08-27 [1] RSPM (R 4.0.5) ## hms 0.5.3 2020-01-08 [1] RSPM (R 4.0.0) ## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.0.2) ## httr 1.4.2 2020-07-20 [1] RSPM (R 4.0.3) ## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.2) ## jsonlite 1.7.1 2020-09-07 [1] RSPM (R 4.0.2) ## knitr 1.33 2024-03-13 [1] Github (yihui/knitr@a1052d1) ## lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.0.2) ## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.0.2) ## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.0.2) ## openssl 1.4.3 2020-09-18 [1] RSPM (R 4.0.3) ## ottrpal 1.2.1 2024-03-13 [1] Github (jhudsl/ottrpal@48e8c44) ## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.0.2) ## pkgbuild 1.1.0 2020-07-13 [1] RSPM (R 4.0.2) ## pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.0.3) ## pkgload 1.1.0 2020-05-29 [1] RSPM (R 4.0.3) ## prettyunits 1.1.1 2020-01-24 [1] RSPM (R 4.0.3) ## processx 3.4.4 2020-09-03 [1] RSPM (R 4.0.2) ## ps 1.4.0 2020-10-07 [1] RSPM (R 4.0.2) ## R6 2.4.1 2019-11-12 [1] RSPM (R 4.0.0) ## readr 1.4.0 2020-10-05 [1] RSPM (R 4.0.2) ## remotes 2.2.0 2020-07-21 [1] RSPM (R 4.0.3) ## rlang 1.1.3 2024-01-10 [1] CRAN (R 4.0.2) ## rmarkdown 2.10 2024-03-13 [1] Github (rstudio/rmarkdown@02d3c25) ## rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.0.2) ## sass 0.4.8 2023-12-06 [1] CRAN (R 4.0.2) ## sessioninfo 1.1.1 2018-11-05 [1] RSPM (R 4.0.3) ## stringi 1.5.3 2020-09-09 [1] RSPM (R 4.0.3) ## stringr 1.4.0 2019-02-10 [1] RSPM (R 4.0.3) ## testthat 3.0.1 2024-03-13 [1] Github (R-lib/testthat@e99155a) ## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.0.2) ## usethis 1.6.3 2020-09-17 [1] RSPM (R 4.0.2) ## utf8 1.1.4 2018-05-24 [1] RSPM (R 4.0.3) ## vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.0.2) ## withr 2.3.0 2020-09-22 [1] RSPM (R 4.0.2) ## xfun 0.26 2024-03-13 [1] Github (yihui/xfun@74c2a66) ## xml2 1.3.2 2020-04-23 [1] RSPM (R 4.0.3) ## yaml 2.2.1 2020-02-01 [1] RSPM (R 4.0.3) ## ## [1] /usr/local/lib/R/site-library ## [2] /usr/local/lib/R/library "],["references-1.html", "Chapter 12 References", " Chapter 12 References "],["404.html", "Page not found", " Page not found The page you requested cannot be found (perhaps it was moved or renamed). You may want to try searching to find the page's new location, or use the table of contents to find the page you are looking for. "]]
+[["index.html", "Intermediate R, Season 3 Chapter 1 About this Course 1.1 Curriculum 1.2 Target Audience 1.3 Offerings", " Intermediate R, Season 3 May, 2024 Chapter 1 About this Course 1.1 Curriculum The course continues building programming fundamentals in R programming and data analysis. You will learn how to make use of complex data structures, use custom functions built by other R users, creating your own functions, and how to iterate repeated tasks that scales naturally. You will also learn how to clean messy data to a Tidy form for analysis, and conduct an end-to-end data science workflow. 1.2 Target Audience The course is intended for researchers who want to continue learning the fundamentals of R programming and how to deal with messy datasets. The audience should know how to subset dataframes and vectors and conduct basic analysis, and/or have taken our Intro to R course. 1.3 Offerings This course is taught on a regular basis at Fred Hutch Cancer Center through the Data Science Lab. Announcements of course offering can be found here. If you wish to follow the course content asynchronously, you may access the course content on this website and exercises and solutions on Posit Cloud. The Posit Cloud compute space can be copied to your own workspace for personal use, or you can access the exercises and solutions on GitHub. "],["fundamentals.html", "Chapter 2 Fundamentals 2.1 Goals of this course 2.2 Data types in R 2.3 Data structures 2.4 Vector 2.5 Factors 2.6 Dataframes 2.7 Lists 2.8 Matrix 2.9 Exercises", " Chapter 2 Fundamentals 2.1 Goals of this course Continue building programming fundamentals: How to use complex data structures, use and create custom functions, and how to iterate repeated tasks Continue exploration of data science fundamentals: how to clean messy data to a Tidy form for analysis. At the end of the course, you will be able to: conduct a full analysis in the data science workflow (minus model). 2.2 Data types in R Numeric: 18, -21, 65, 1.25 Character: “ATCG”, “Whatever”, “948-293-0000” Logical: TRUE, FALSE Missing values: NA 2.3 Data structures Vector Factor Dataframe List Matrix 2.4 Vector We know what an (atomic) vector is: it can contains a data type, and all elements must be the same data type. Within the Numeric type that we are familiar with, there are more specific types: Integer consists of whole number values, and Double consists of decimal values. Most of the time we only need to consider Numeric types, but once in a while we need to be more specific. We can test whether a vector is a certain type with is.___() functions, such as is.character(). is.character(c("hello", "there")) ## [1] TRUE For NA, the test will return a vector testing each element, because NA can be mixed into other values: is.na(c(34, NA)) ## [1] FALSE TRUE We can coerce vectors from one type to the other with as.___() functions, such as as.numeric() as.numeric(c("23", "45")) ## [1] 23 45 as.numeric(c(TRUE, FALSE)) ## [1] 1 0 It is common to have metadata attributes, such as names, attached to R data structures. x = c(1, 2, 3) names(x) = c("a", "b", "c") x ## a b c ## 1 2 3 We can look for more general attributes via the attributes() function: attributes(x) ## $names ## [1] "a" "b" "c" 2.4.1 Ways to subset a vector Positive numeric vector Negative numeric vector performs exclusion Logical vector 2.4.2 Practice implicit subsetting How do you subset the following vector so that it only has positive values? data = c(2, 4, -1, -3, 2, -1, 10) data[data > 0] ## [1] 2 4 2 10 How do you subset the following vector so that it has doesn’t have the character “temp”? chars = c("temp", "object", "temp", "wish", "bumblebee", "temp") chars[chars != "temp"] ## [1] "object" "wish" "bumblebee" How do you subset the following vector so that it has no NA values? vec_with_NA = c(2, 4, NA, NA, 3, NA) vec_with_NA[!is.na(vec_with_NA)] ## [1] 2 4 3 2.5 Factors Factors are a type of vector that holds categorical information, such as sex, gender, or cancer subtype. They are useful for: When you know you have a fixed number of categories. When you want to display character vectors in a non-alphabetical order, which is common in plotting. Inputs for statistical models, as factors are a special type of numerical vectors. place = factor(c("first", "third", "third", "second", "second", "fourth")) place ## [1] first third third second second fourth ## Levels: first fourth second third df = data.frame(p = place) ggplot(df) + geom_bar(aes(x = p)) We can construct ordered factors: place = ordered(c("first", "third", "third", "second","second", "fourth"), levels = c("first", "second", "third", "fourth")) place ## [1] first third third second second fourth ## Levels: first < second < third < fourth df = data.frame(p = place) ggplot(df) + geom_bar(aes(x = p)) 2.6 Dataframes Usually, we load in a dataframe from a spreadsheet or a package, but we can create a new dataframe by using vectors of the same length via the data.frame() function: df = data.frame(x = 1:3, y = c("cup", "mug", "jar")) attributes(df) ## $names ## [1] "x" "y" ## ## $class ## [1] "data.frame" ## ## $row.names ## [1] 1 2 3 library(palmerpenguins) attributes(penguins) ## $class ## [1] "tbl_df" "tbl" "data.frame" ## ## $row.names ## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 ## [19] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 ## [37] 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 ## [55] 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 ## [73] 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 ## [91] 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 ## [109] 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 ## [127] 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 ## [145] 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 ## [163] 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 ## [181] 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 ## [199] 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 ## [217] 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 ## [235] 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 ## [253] 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 ## [271] 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 ## [289] 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 ## [307] 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 ## [325] 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 ## [343] 343 344 ## ## $names ## [1] "species" "island" "bill_length_mm" ## [4] "bill_depth_mm" "flipper_length_mm" "body_mass_g" ## [7] "sex" "year" Why are row names undesirable? Sometimes, data frames will be in a format called “tibble”, as shown in the penguins class names as “tbl_df”, and “tbl”. 2.6.1 Subsetting dataframes Getting one single column: penguins$bill_length_mm ## [1] 39.1 39.5 40.3 NA 36.7 39.3 38.9 39.2 34.1 42.0 37.8 37.8 41.1 38.6 34.6 ## [16] 36.6 38.7 42.5 34.4 46.0 37.8 37.7 35.9 38.2 38.8 35.3 40.6 40.5 37.9 40.5 ## [31] 39.5 37.2 39.5 40.9 36.4 39.2 38.8 42.2 37.6 39.8 36.5 40.8 36.0 44.1 37.0 ## [46] 39.6 41.1 37.5 36.0 42.3 39.6 40.1 35.0 42.0 34.5 41.4 39.0 40.6 36.5 37.6 ## [61] 35.7 41.3 37.6 41.1 36.4 41.6 35.5 41.1 35.9 41.8 33.5 39.7 39.6 45.8 35.5 ## [76] 42.8 40.9 37.2 36.2 42.1 34.6 42.9 36.7 35.1 37.3 41.3 36.3 36.9 38.3 38.9 ## [91] 35.7 41.1 34.0 39.6 36.2 40.8 38.1 40.3 33.1 43.2 35.0 41.0 37.7 37.8 37.9 ## [106] 39.7 38.6 38.2 38.1 43.2 38.1 45.6 39.7 42.2 39.6 42.7 38.6 37.3 35.7 41.1 ## [121] 36.2 37.7 40.2 41.4 35.2 40.6 38.8 41.5 39.0 44.1 38.5 43.1 36.8 37.5 38.1 ## [136] 41.1 35.6 40.2 37.0 39.7 40.2 40.6 32.1 40.7 37.3 39.0 39.2 36.6 36.0 37.8 ## [151] 36.0 41.5 46.1 50.0 48.7 50.0 47.6 46.5 45.4 46.7 43.3 46.8 40.9 49.0 45.5 ## [166] 48.4 45.8 49.3 42.0 49.2 46.2 48.7 50.2 45.1 46.5 46.3 42.9 46.1 44.5 47.8 ## [181] 48.2 50.0 47.3 42.8 45.1 59.6 49.1 48.4 42.6 44.4 44.0 48.7 42.7 49.6 45.3 ## [196] 49.6 50.5 43.6 45.5 50.5 44.9 45.2 46.6 48.5 45.1 50.1 46.5 45.0 43.8 45.5 ## [211] 43.2 50.4 45.3 46.2 45.7 54.3 45.8 49.8 46.2 49.5 43.5 50.7 47.7 46.4 48.2 ## [226] 46.5 46.4 48.6 47.5 51.1 45.2 45.2 49.1 52.5 47.4 50.0 44.9 50.8 43.4 51.3 ## [241] 47.5 52.1 47.5 52.2 45.5 49.5 44.5 50.8 49.4 46.9 48.4 51.1 48.5 55.9 47.2 ## [256] 49.1 47.3 46.8 41.7 53.4 43.3 48.1 50.5 49.8 43.5 51.5 46.2 55.1 44.5 48.8 ## [271] 47.2 NA 46.8 50.4 45.2 49.9 46.5 50.0 51.3 45.4 52.7 45.2 46.1 51.3 46.0 ## [286] 51.3 46.6 51.7 47.0 52.0 45.9 50.5 50.3 58.0 46.4 49.2 42.4 48.5 43.2 50.6 ## [301] 46.7 52.0 50.5 49.5 46.4 52.8 40.9 54.2 42.5 51.0 49.7 47.5 47.6 52.0 46.9 ## [316] 53.5 49.0 46.2 50.9 45.5 50.9 50.8 50.1 49.0 51.5 49.8 48.1 51.4 45.7 50.7 ## [331] 42.5 52.2 45.2 49.3 50.2 45.6 51.9 46.8 45.7 55.8 43.5 49.6 50.8 50.2 I want to select columns bill_length_mm, bill_depth_mm, species, and filter for species that are “Gentoo”: penguins_select = select(penguins, bill_length_mm, bill_depth_mm, species) penguins_gentoo = filter(penguins_select, species == "Gentoo") or penguins_select_2 = penguins[, c("bill_length_mm", "bill_depth_mm", "species")] penguins_gentoo_2 = penguins_select_2[penguins$species == "Gentoo" ,] or penguins_gentoo_2 = penguins_select_2[penguins$species == "Gentoo", c("bill_length_mm", "bill_depth_mm", "species")] I want to filter out rows that have NAs in the column bill_length_mm: penguins_clean = filter(penguins, !is.na(bill_length_mm)) or penguins_clean = penguins[!is.na(penguins$bill_depth_mm) ,] 2.7 Lists Lists operate similarly as vectors as they group data into one dimension, but each element of a list can be any data type or data structure! l1 = list( 1:3, "a", c(TRUE, FALSE, TRUE), c(2.3, 5.9) ) Unlike vectors, you access the elements of a list via the double bracket [[]]. You access a smaller list with single bracket []. (More discussion on the different uses of the bracket here.) Here’s a nice metaphor: If list x is a train carrying objects, then x[[5]] is the object in car 5; x[4:6] is a train of cars 4-6. l1[[1]] ## [1] 1 2 3 l1[[1]][2] ## [1] 2 Use unlist() to coerce a list into a vector. Notice all the automatic coersion that happened for the elements. unlist(l1) ## [1] "1" "2" "3" "a" "TRUE" "FALSE" "TRUE" "2.3" "5.9" We can give names to lists: l1 = list( ranking = 1:3, name = "a", success = c(TRUE, FALSE, TRUE), score = c(2.3, 5.9) ) #or names(l1) = c("ranking", "name", "success", "score") And access named elements of lists via the $ operation: l1$score ## [1] 2.3 5.9 Therefore, l1$score is the same as l1[[4]] and is the same as l1[[\"score\"]]. A dataframe is just a named list of vectors of same length with additional attributes of (column) names and row.names. 2.8 Matrix A matrix holds information of the same data type in two dimensions - it’s like a two dimensional vector. Matricies are most often used in statistical computing and matrix algebra, such as creating a design matrix. They are often created by taking a vector and reshaping it with a set number of rows and columns, or converting from a dataframe with only one data type. my_matrix = matrix(1:10, nrow = 2) my_matrix ## [,1] [,2] [,3] [,4] [,5] ## [1,] 1 3 5 7 9 ## [2,] 2 4 6 8 10 You access elements of a matrix similar to that of a dataframe’s indexing: #column 3 my_matrix[, 3] ## [1] 5 6 #row 2 my_matrix[2 ,] ## [1] 2 4 6 8 10 #column 3, row 2 my_matrix[2, 3] ## [1] 6 2.9 Exercises You can find exercises and solutions on Posit Cloud, or on GitHub. "],["data-cleaning-part-1.html", "Chapter 3 Data Cleaning, Part 1 3.1 Interpreting functions, carefully 3.2 Recoding Data / Conditionals 3.3 Conditionals 3.4 Exercises", " Chapter 3 Data Cleaning, Part 1 3.1 Interpreting functions, carefully As you become more independent R programmers, you will spend time learning about new functions on your own. We have gone over the basic anatomy of a function call back in Intro to R, but now let’s go a bit deeper to understand how a function is built and how to call them. Recall that a function has a function name, input arguments, and a return value. Function definition consists of assigning a function name with a “function” statement that has a comma-separated list of named function arguments, and a return expression. The function name is stored as a variable in the global environment. In order to use the function, one defines or import it, then one calls it. Example: addFunction = function(num1, num2) { result = num1 + num2 return(result) } result = addFunction(3, 4) When the function is called in line 5, the variables for the arguments are reassigned to function arguments to be used within the function and helps with the modular form. What do you think are some valid inputs for this function? To see why we need the variables of the arguments to be reassigned, consider the following function that is not modular: x = 3 y = 4 addFunction = function(num1, num2) { result = x + y return(result) } result = addFunction(10, -10) Some syntax equivalents on calling the function: addFunction(3, 4) addFunction(num1 = 3, num2 = 4) addFunction(num2 = 4, num1 = 3) but this could be different: addFunction(4, 3) With a deeper knowledge of how functions are built, when you encounter a foreign function, you can look up its help page to understand how to use it. For example, let’s look at mean(): ?mean Arithmetic Mean Description: Generic function for the (trimmed) arithmetic mean. Usage: mean(x, ...) ## Default S3 method: mean(x, trim = 0, na.rm = FALSE, ...) Arguments: x: An R object. Currently there are methods for numeric/logical vectors and date, date-time and time interval objects. Complex vectors are allowed for ‘trim = 0’, only. trim: the fraction (0 to 0.5) of observations to be trimmed from each end of ‘x’ before the mean is computed. Values of trim outside that range are taken as the nearest endpoint. na.rm: a logical evaluating to ‘TRUE’ or ‘FALSE’ indicating whether ‘NA’ values should be stripped before the computation proceeds. ...: further arguments passed to or from other methods. Notice that the arguments trim = 0, na.rm = FALSE have default values. This means that these arguments are optional - you should provide it only if you want to. With this understanding, you can use mean() in a new way: numbers = c(1, 2, NA, 4) mean(x = numbers, na.rm = TRUE) ## [1] 2.333333 The use of . . . (dot-dot-dot): This is a special argument that allows a function to take any number of arguments. This isn’t very useful for the mean() function, but it makes sense for function such as select() and filter(), and mutate(). For instance, in select(), once you provide your dataframe for the argument .data, you can pile on as many columns to select in the rest of the argument. Usage: select(.data, ...) Arguments: .data: A data frame, data frame extension (e.g. a tibble), or a lazy data frame (e.g. from dbplyr or dtplyr). See _Methods_, below, for more details. ...: <‘tidy-select’> One or more unquoted expressions separated by commas. Variable names can be used as if they were positions in the data frame, so expressions like ‘x:y’ can be used to select a range of variables. You will look at the function documentation on your own to see how to deal with more complex cases. 3.2 Recoding Data / Conditionals It is often said that 80% of data analysis is spent on the cleaning and preparing data. Today we will start looking at common data cleaning tasks. Suppose that you have a column in your data that needs to be recoded. Since a dataframe’s column, when selected via $, is a vector, let’s start talking about recoding vectors. If we have a numeric vector, then maybe you want to have certain values to be out of bounds, or assign a range of values to a character category. If we have a character vector, then maybe you want to reassign it to a different value. Here are popular recoding logical scenarios: If: “If elements of the vector meets condition, then they are assigned value.” If-else: “If elements of the vector meets condition, then they are assigned value X. Otherwise, they are assigned value Y.” If-else_if-else: “If elements of the vector meets condition A, then they are assigned value X. Else, if the elements of the vector meets condition B, they are assigned value Y. Otherwise, they are assigned value Z.” Let’s look at a vector of grade values, as an example: grade = c(90, 78, 95, 74, 56, 81, 102) If Instead of having the bracket [ ] notation on the right hand side of the equation, if it is on the left hand side of the equation, then we can modify a subset of the vector. grade1 = grade grade1[grade1 > 100] = 100 If-else grade2 = if_else(grade > 60, TRUE, FALSE) If-else_if-else grade3 = case_when(grade >= 90 ~ "A", grade >= 80 ~ "B", grade >= 70 ~ "C", grade >= 60 ~ "D", .default = "F") Let’s do it for dataframes now. simple_df = data.frame(grade = c(90, 78, 95, 74, 56, 81, 102), status = c("case", " ", "Control", "control", "Control", "Case", "case")) If simple_df1 = simple_df simple_df1$grade[simple_df1$grade > 100] = 100 If-else simple_df2 = simple_df simple_df2$grade = ifelse(simple_df2$grade > 60, TRUE, FALSE) or simple_df2 = mutate(simple_df, grade = ifelse(grade > 60, TRUE, FALSE)) If-else_if-else simple_df3 = simple_df simple_df3$grade = case_when(simple_df3$grade >= 90 ~ "A", simple_df3$grade >= 80 ~ "B", simple_df3$grade >= 70 ~ "C", simple_df3$grade >= 60 ~ "D", .default = "F") or simple_df3 = simple_df simple_df3 = mutate(simple_df3, grade = case_when(grade >= 90 ~ "A", grade >= 80 ~ "B", grade >= 70 ~ "C", grade >= 60 ~ "D", .default = "F")) 3.3 Conditionals The 3 common scenarios we looked at for recoding data is closely tied to the concept of conditionals in programming: given certain conditions, you run a specific code chunk. Given a vector’s value, assign it a different value. Or, given a value, run the following hundred lines of code. Here is what it looks like: If: if(expression_is_TRUE) { #code goes here } If-else: if(expression_is_TRUE) { #code goes here }else { #other code goes here } If-else_if-else: if(expression_A_is_TRUE) { #code goes here }else if(expression_B_is_TRUE) { #other code goes here }else { #some other code goes here } The expression that is being tested whether it is TRUE must be a singular logical value, and not a logical vector. If the latter, see the recoding section for now. Example: nuc = "A" if(nuc == "A") { nuc = "T" }else if(nuc == "T") { nuc = "A" }else if(nuc == "C") { nuc = "C" }else if(nuc == "G") { nuc = "C" }else { nuc = NA } nuc ## [1] "T" Example: my_input = c(1, 3, 5, 7, 9) #my_input = c("e", "e", "a", "i", "o") if(is.numeric(my_input)) { result = mean(my_input) }else if(is.character(my_input)) { result = table(my_input) } result ## [1] 5 3.4 Exercises You can find exercises and solutions on Posit Cloud, or on GitHub. "],["data-cleaning-part-2.html", "Chapter 4 Data Cleaning, Part 2 4.1 Tidy Data 4.2 Uses of Tidy data 4.3 Subjectivity in Tidy Data 4.4 Exercises", " Chapter 4 Data Cleaning, Part 2 library(tidyverse) 4.1 Tidy Data It is important to have standard of organizing data, as it facilitates a consistent way of thinking about data organization and building tools (functions) that make use of that standard. The principles of Tidy data, developed by Hadley Wickham: Each variable must have its own column. Each observation must have its own row. Each value must have its own cell. If we want to be technical about what variables and observations are, Hadley Wickham describes: A dataset is a collection of values, usually either numbers (if quantitative) or strings (if qualitative). Every value belongs to a variable and an observation. A variable contains all values that measure the same underlying attribute (like height, temperature, duration) across units. An observation contains all values measured on the same unit (like a person, or a day, or a race) across attributes. A tidy dataframe. Besides a standard, Tidy data is useful because many tools in R are most effective when your data is in a Tidy format. This includes data visualization with ggplot, regression models, databases, and more. These tools assumes the values of each variable fall in their own column vector. It seems hard to go wrong with these simple criteria of Tidy data! However, in reality, many dataframes we load in aren’t Tidy, and it’s easiest seen through counterexamples and how to fix it. Here are some common ways that data becomes un-Tidy: Columns contain values of variables, rather than variables Variables are stored in rows Multiple variables are stored in a single column After some clear examples, we emphasize that “Tidy” data is subjective to what kind of analysis you want to do with the dataframe. 4.1.1 1. Columns contain values, rather than variables (Long is tidy) df = data.frame(Store = c("A", "B"), Year = c(2018, 2018), Q1_Sales = c(55, 98), Q2_Sales = c(45, 70), Q3_Sales = c(22, 60), Q4_Sales = c(50, 60)) df ## Store Year Q1_Sales Q2_Sales Q3_Sales Q4_Sales ## 1 A 2018 55 45 22 50 ## 2 B 2018 98 70 60 60 Each observation is a store, and each observation has its own row. That looks good. The columns “Q1_Sales”, …, “Q4_Sales” seem to be values of a single variable “quarter” of our observation. The values of “quarter” are not in a single column, but are instead in the columns. df_long = pivot_longer(df, c("Q1_Sales", "Q2_Sales", "Q3_Sales", "Q4_Sales"), names_to = "quarter", values_to = "sales") df_long ## # A tibble: 8 × 4 ## Store Year quarter sales ## <chr> <dbl> <chr> <dbl> ## 1 A 2018 Q1_Sales 55 ## 2 A 2018 Q2_Sales 45 ## 3 A 2018 Q3_Sales 22 ## 4 A 2018 Q4_Sales 50 ## 5 B 2018 Q1_Sales 98 ## 6 B 2018 Q2_Sales 70 ## 7 B 2018 Q3_Sales 60 ## 8 B 2018 Q4_Sales 60 Now, each observation is a store’s quarter, and each observation has its own row. The new columns “quarter” and “sales” are variables that describes our observation, and describes our values. We’re in a tidy state! We have transformed our data to a “longer” format, as our observation represents something more granular or detailed than before. Often, the original variables values will repeat itself in a “longer format”. We call the previous state of our dataframe is a “wider” format. 4.1.2 2. Variables are stored in rows (Wide is tidy) Are all tidy dataframes Tidy in a “longer” format? df2 = data.frame(Sample = c("A", "B"), KRAS_mutation = c(TRUE, FALSE), KRAS_expression = c(2.3, 3.9)) df2 ## Sample KRAS_mutation KRAS_expression ## 1 A TRUE 2.3 ## 2 B FALSE 3.9 Each observation is a sample, and each observation has its own row. Looks good. Each variable has its own column, and no values are in columns. What happens if we make it longer? df2_long = pivot_longer(df2, c("KRAS_mutation", "KRAS_expression"), names_to = "gene", values_to = "values") df2_long ## # A tibble: 4 × 3 ## Sample gene values ## <chr> <chr> <dbl> ## 1 A KRAS_mutation 1 ## 2 A KRAS_expression 2.3 ## 3 B KRAS_mutation 0 ## 4 B KRAS_expression 3.9 Here, each observation is a sample’s gene…type? The observation feels awkward because variables are stored in rows. Also, the column “values” contains multiple variable types: gene expression and mutation values that got coerced to numeric! To make this dataframe wider, df2_long_wide = pivot_wider(df2_long, names_from = "gene", values_from = "values") df2_long_wide$KRAS_mutation = as.logical(df2_long_wide$KRAS_mutation) df2_long_wide ## # A tibble: 2 × 3 ## Sample KRAS_mutation KRAS_expression ## <chr> <lgl> <dbl> ## 1 A TRUE 2.3 ## 2 B FALSE 3.9 We are back to our orignal form, and it was already Tidy. 4.1.3 3. Multiple variables are stored in a single column table3 ## # A tibble: 6 × 3 ## country year rate ## * <chr> <int> <chr> ## 1 Afghanistan 1999 745/19987071 ## 2 Afghanistan 2000 2666/20595360 ## 3 Brazil 1999 37737/172006362 ## 4 Brazil 2000 80488/174504898 ## 5 China 1999 212258/1272915272 ## 6 China 2000 213766/1280428583 There seems to be two variables in the numerator and denominator of “rate” column. Let’s separate it. separate(table3, col = "rate", into = c("count", "population"), sep = "/") ## # A tibble: 6 × 4 ## country year count population ## <chr> <int> <chr> <chr> ## 1 Afghanistan 1999 745 19987071 ## 2 Afghanistan 2000 2666 20595360 ## 3 Brazil 1999 37737 172006362 ## 4 Brazil 2000 80488 174504898 ## 5 China 1999 212258 1272915272 ## 6 China 2000 213766 1280428583 4.2 Uses of Tidy data In general, many functions for analysis and visualization in R assumes that the input dataframe is Tidy. These tools assumes the values of each variable fall in their own column vector. For instance, from our first example, we can compare sales across quarters and stores. df_long ## # A tibble: 8 × 4 ## Store Year quarter sales ## <chr> <dbl> <chr> <dbl> ## 1 A 2018 Q1_Sales 55 ## 2 A 2018 Q2_Sales 45 ## 3 A 2018 Q3_Sales 22 ## 4 A 2018 Q4_Sales 50 ## 5 B 2018 Q1_Sales 98 ## 6 B 2018 Q2_Sales 70 ## 7 B 2018 Q3_Sales 60 ## 8 B 2018 Q4_Sales 60 ggplot(df_long) + aes(x = quarter, y = sales, group = Store) + geom_point() + geom_line() Although in its original state we can also look at sales between quarter, we can only look between two quarters at once. Tidy data encourages looking at data in the most granular scale. ggplot(df) + aes(x = Q1_Sales, y = Q2_Sales, color = Store) + geom_point() 4.3 Subjectivity in Tidy Data We have looked at clear cases of when a dataset is Tidy. In reality, the Tidy state depends on what we call variables and observations. Consider this example, inspired by the following blog post by Damien Martin. kidney = data.frame(stone_size = c("Small", "Large"), treatment.A_recovered = c(81, 192), treatment.A_failed = c(6, 71), treatment.B_recovered = c(234, 55), treatment.B_failed = c(36, 25)) kidney ## stone_size treatment.A_recovered treatment.A_failed treatment.B_recovered ## 1 Small 81 6 234 ## 2 Large 192 71 55 ## treatment.B_failed ## 1 36 ## 2 25 Right now, the kidney dataframe clearly has values of a variable in the column. Let’s try to make it Tidy by making it into a longer form and separating out variables that are together in a column. kidney_long = pivot_longer(kidney, c("treatment.A_recovered", "treatment.A_failed", "treatment.B_recovered", "treatment.B_failed"), names_to = "treatment_outcome", values_to = "count") kidney_long = separate(kidney_long, "treatment_outcome", c("treatment", "outcome"), "_") kidney_long ## # A tibble: 8 × 4 ## stone_size treatment outcome count ## <chr> <chr> <chr> <dbl> ## 1 Small treatment.A recovered 81 ## 2 Small treatment.A failed 6 ## 3 Small treatment.B recovered 234 ## 4 Small treatment.B failed 36 ## 5 Large treatment.A recovered 192 ## 6 Large treatment.A failed 71 ## 7 Large treatment.B recovered 55 ## 8 Large treatment.B failed 25 Here, each observation is a kidney stone’s treatment’s outcome type, and each observation has its own row. The column “count” describes our observation, and describes our values. This dataframe seems reasonably Tidy. How about this? kidney_long_still = pivot_wider(kidney_long, names_from = "outcome", values_from = "count") kidney_long_still ## # A tibble: 4 × 4 ## stone_size treatment recovered failed ## <chr> <chr> <dbl> <dbl> ## 1 Small treatment.A 81 6 ## 2 Small treatment.B 234 36 ## 3 Large treatment.A 192 71 ## 4 Large treatment.B 55 25 Here, each observation is a kidney stone’s treatment, and each observation has its own row. The columns “recovered” and “failed” are variables that describes our observation, and describes its corresponding values. This dataframe seems reasonably Tidy, also. The reason why both of these versions seem Tidy is that the columns “recovered” and “failed” can be interpreted as independent variables and values of the variable “treatment”. Ultimately, we decide which dataframe we prefer based on the analysis we want to do. For instance, when our observation is about a kidney stone’s treatment’s outcome type, we compare it between outcome type, treatment, and stone size. ggplot(kidney_long) + aes(x = treatment, y = count, fill = outcome) + geom_bar(position="dodge", stat="identity") + facet_wrap(~stone_size) When our observation is about a kidney stone’s treatment’s, we compare a new variable recovery rate ( = recovered / (recovered + failed)) between treatment and stone size. kidney_long_still = mutate(kidney_long_still, recovery_rate = recovered / (recovered + failed)) ggplot(kidney_long_still) + aes(x = treatment, y = recovery_rate, fill = stone_size) + geom_bar(position="dodge", stat="identity") 4.4 Exercises You can find exercises and solutions on Posit Cloud, or on GitHub. "],["writing-your-first-function.html", "Chapter 5 Writing your first function 5.1 Anatomy of a function definition 5.2 Local and global environments 5.3 A step-by-step example 5.4 Function arguments create modularity 5.5 Examples 5.6 Exercises", " Chapter 5 Writing your first function Function machine from algebra class. We write functions for two main, often overlapping, reasons: Following DRY (Don’t Repeat Yourself) principle: If you find yourself repeating similar patterns of code, you should write a function that executes that pattern. This saves time and the risk of mistakes. Create modular structure and abstraction: Having all of your code in one place becomes increasingly complicated as your program grows. Think of the function as a mini-program that can perform without the rest of the program. Organizing your code by functions gives modular structure, as well as abstraction: you only need to know the function name, inputs, and output to use it and don’t have to worry how it works. Some advice on writing functions: Code that has a well-defined set of inputs and outputs make a good function. A function should do only one, well-defined task. 5.1 Anatomy of a function definition Function definition consists of assigning a function name with a “function” statement that has a comma-separated list of named function arguments, and a return expression. The function name is stored as a variable in the global environment. In order to use the function, one defines or import it, then one calls it. Example: addFunction = function(argument1, argument2) { result = argument1 + argument2 return(result) } z = addFunction(3, 4) With function definitions, not all code runs from top to bottom. The first four lines defines the function, but the function is never run. It is called on line 5, and the lines within the function are executed. When the function is called in line 5, the variables for the arguments are reassigned to function arguments to be used within the function and helps with the modular form. We need to introduce the concept of local and global environments to distinguish variables used only for a function from variables used for the entire program. 5.2 Local and global environments { } represents variable scoping: within each { }, if variables are defined, they are stored in a local environment, and is only accessible within { }. All function arguments are stored in the local environment. The overall environment of the program is called the global environment and can be also accessed within { }. The reason of having some of this “privacy” in the local environment is to make functions modular - they are independent little tools that should not interact with the rest of the global environment. Imagine someone writing a tool that they want to give someone else to use, but the tool depends on your environment, vice versa. 5.3 A step-by-step example Using the addFunction function, let’s see step-by-step how the R interpreter understands our code: We define the function in the global environment. We call the function, and the function arguments 3, 4 are assigned to argument1 and argument2, respectively in the function’s local environment. We run the first line of code in the function body. The new variable “result” is stored in the local environment because it is within { }. We run the second line of code in the function body to return a value. The return value from the function is assigned to the variable z in the global environment. All local variables for the function are erased now that the function call is over. 5.4 Function arguments create modularity First time writers of functions might ask: why are variables we use for the arguments of a function reassigned for function arguments in the local environment? Here is an example when that process is skipped - what are the consequences? x = 3 y = 4 addFunction = function(argument1, argument2) { result = x + y return(result) } z = addFunction(x, y) w = addFunction(10, -5) What do you expect the value of z to be? How about w? Here is the execution for w: We define the variables and function in the global environment. We call the function, and the function arguments 10, -5 are assigned to argument1 and argument2, respectively in the function’s local environment. We run the first line of code in the function body. The new variable “result” is stored in the local environment because it is within { }. We run the second line of code in the function body to return a value. The return value from the function is assigned to the variable w in the global environment. All local variables for the function are erased now that the function call is over. The function did not work as expected because we used hard-coded variables from the global environment and not function argument variables unique to the function use! 5.5 Examples Create a function, called add_and_raise_power in which the function takes in 3 numeric arguments. The function computes the following: the first two arguments are added together and raised to a power determined by the 3rd argument. The function returns the resulting value. Here is a use case: add_and_raise_power(1, 2, 3) = 27 because the function will return this expression: (1 + 2) ^ 3. Another use case: add_and_raise_power(3, 1, 2) = 16 because of the expression (3 + 1) ^ 2. Confirm with that these use cases work. Can this function used for numeric vectors? add_and_raise_power = function(x, y, z) { result = (x + y)^z return(result) } add_and_raise_power(1, 2, 3) ## [1] 27 Create a function, called my_dim in which the function takes in one argument: a dataframe. The function returns the following: a length-2 numeric vector in which the first element is the number of rows in the dataframe, and the second element is the number of columns in the dataframe. Your result should be identical as the dim function. How can you leverage existing functions such as nrow and ncol? Use case: my_dim(penguins) = c(344, 8) library(palmerpenguins) my_dim = function(df) { result = c(nrow(df), ncol(df)) return(result) } my_dim(penguins) ## [1] 344 8 Create a function, called num_na in which the function takes in any vector, and then return a single numeric value. This numeric value is the number of NAs in the vector. Use cases: num_na(c(NA, 2, 3, 4, NA, 5)) = 2 and num_na(c(2, 3, 4, 5)) = 0. Hint 1: Use is.na() function. Hint 2: Given a logical vector, you can count the number of TRUE values by using sum(), such as sum(c(TRUE, TRUE, FALSE)) = 2. num_na = function(x) { return(sum(is.na(num_na))) } Create a function, called medicaid_eligible in which the function takes in one argument: a numeric vector called age. The function returns a numeric vector with the same length as age, in which elements are 0 for indicies that are less than 65 in age, and 1 for indicies 65 or higher in age. (Hint: This is a data recoding problem!) Use cases: medicaid_eligible(c(30, 70)) = c(0, 1) medicaid_eligible = function(age) { result = age result[age < 65] = 0 result[age >= 65] = 1 return(result) } medicaid_eligible(c(30, 70)) ## [1] 0 1 5.6 Exercises You can find exercises and solutions on Posit Cloud, or on GitHub. "],["iteration.html", "Chapter 6 Iteration 6.1 For loops 6.2 Functionals 6.3 Case studies 6.4 Exercises", " Chapter 6 Iteration Suppose that you want to repeat a chunk of code many times, but changing one variable’s value each time you do it. This could be modifying each element of a vector with the same operation, or analyzing a dataframe with different parameters. There are three common strategies to go about this: Copy and paste the code chunk, and change that variable’s value. Repeat. This can be a starting point in your analysis, but will lead to errors easily. Use a for loop to repeat the chunk of code, and let it loop over the changing variable’s value. This is popular for many programming languages, but the R programming culture encourages a functional way instead. Functionals allow you to take a function that solves the problem for a single input and generalize it to handle any number of inputs. This is very popular in R programming culture. 6.1 For loops A for loop repeats a chunk of code many times, once for each element of an input vector. for (my_element in my_vector) { chunk of code } Most often, the “chunk of code” will make use of my_element. 6.1.0.1 We can loop through the indicies of a vector The function seq_along() creates the indicies of a vector. It has almost the same properties as 1:length(my_vector), but avoids issues when the vector length is 0. my_vector = c(1, 3, 5, 7) for(i in seq_along(my_vector)) { print(my_vector[i]) } ## [1] 1 ## [1] 3 ## [1] 5 ## [1] 7 6.1.0.2 Alternatively, we can loop through the elements of a vector for(vec_i in my_vector) { print(vec_i) } ## [1] 1 ## [1] 3 ## [1] 5 ## [1] 7 6.1.0.3 Another example via indicies result = rep(NA, length(my_vector)) for(i in seq_along(my_vector)) { result[i] = log(my_vector[i]) } 6.2 Functionals A functional is a function that takes in a data structure and function as inputs and applies the function on the data structure, element by element. It maps your input data structure to an output data structure based on the function. It encourages the usage of modular functions in your code. Or, We will use the purrr package in tidyverse to use functionals. map() takes in a vector or a list, and then applies the function on each element of it. The output is always a list. my_vector = c(1, 3, 5, 7) map(my_vector, log) ## [[1]] ## [1] 0 ## ## [[2]] ## [1] 1.098612 ## ## [[3]] ## [1] 1.609438 ## ## [[4]] ## [1] 1.94591 Lists are useful if what you are using it on requires a flexible data structure. To be more specific about the output type, you can do this via the map_* function, where * specifies the output type: map_lgl(), map_chr(), and map_dbl() functions return vectors of logical values, strings, or numbers respectively. For example, to make sure your output is a double (numeric): map_dbl(my_vector, log) ## [1] 0.000000 1.098612 1.609438 1.945910 All of these are toy examples that gets us familiar with the syntax, but we already have built-in functions to solve these problems, such as log(my_vector). Let’s see some real-life case studies. 6.3 Case studies 6.3.1 1. Loading in multiple files. Suppose that we want to load in a few dataframes, and store them in a list of dataframes for analysis downstream. We start with the filepaths we want to load in as dataframes. paths = c("classroom_data/students.csv", "classroom_data/CCLE_metadata.csv") The function we want to use to load the data in will be read_csv(). Let’s practice writing out one iteration: result = read_csv(paths[1]) 6.3.1.1 To do this functionally, we think about: What variable we need to loop through: paths The repeated task as a function: read_csv() The looping mechanism, and its output: map() outputs lists. loaded_dfs = map(paths, read_csv) 6.3.1.2 To do this with a for loop, we think about: What variable we need to loop through: paths. Do we need to store the outcome of this loop in a data structure? Yes, a list. At each iteration, what are we doing? Use read_csv() on the current element, and store it in the output list. paths = c("classroom_data/students.csv", "classroom_data/CCLE_metadata.csv") loaded_dfs = vector(mode = "list", length = length(paths)) for(i in seq_along(paths)) { df = read_csv(paths[i]) loaded_dfs[[i]] = df } 6.3.2 2. Analyze a dataframe with different parameters. Suppose you are working with the penguins dataframe: library(palmerpenguins) head(penguins) ## # A tibble: 6 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## # ℹ 2 more variables: sex <fct>, year <int> and you want to look at the mean bill_length_mm for each of the three species (Adelie, Chinstrap, Gentoo). Let’s practice writing out one iteration: species_to_analyze = c("Adelie", "Chinstrap", "Gentoo") penguins_subset = filter(penguins, species == species_to_analyze[1]) mean(penguins_subset$bill_length_mm, na.rm = TRUE) ## [1] 38.79139 6.3.2.1 To do this functionally, we think about: What variable we need to loop through: c(\"Adelie\", \"Chinstrap\", \"Gentoo\") The repeated task as a function: a custom function that takes in a specie of interest. The function filters the rows of penguins to the species of interest, and compute the mean of bill_length_mm. The looping mechanism, and its output: map_dbl() outputs (double) numeric vectors. analysis = function(current_species) { penguins_subset = dplyr::filter(penguins, species == current_species) return(mean(penguins_subset$bill_length_mm, na.rm=TRUE)) } map_dbl(c("Adelie", "Chinstrap", "Gentoo"), analysis) ## [1] 38.79139 48.83382 47.50488 6.3.2.2 To do this with a for loop, we think about: What variable we need to loop through: c(\"Adelie\", \"Chinstrap\", \"Gentoo\"). Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector. At each iteration, what are we doing? Filter the rows of penguins to the species of interest, and compute the mean of bill_length_mm. outcome = rep(NA, length(species_to_analyze)) for(i in seq_along(species_to_analyze)) { penguins_subset = filter(penguins, species == species_to_analyze[i]) outcome[i] = mean(penguins_subset$bill_length_mm, na.rm=TRUE) } outcome ## [1] 38.79139 48.83382 47.50488 6.3.3 3. Calculate summary statistics on columns of a dataframe. Suppose that you are interested in the numeric columns of the penguins dataframe. penguins_numeric = penguins %>% select(bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g) and you are interested to look at the mean of each column. It is very helpful to interpret the dataframe penguins_numeric as a list, iterating through each column as an element of a list. Let’s practice writing out one iteration: mean(penguins_numeric[[1]], na.rm = TRUE) ## [1] 43.92193 6.3.3.1 To do this functionally, we think about: What variable we need to loop through: the list penguins_numeric The repeated task as a function: mean() with the argument na.rm = TRUE. The looping mechanism, and its output: map_dbl() outputs (double) numeric vectors. map_dbl(penguins_numeric, mean, na.rm = TRUE) ## bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## 43.92193 17.15117 200.91520 4201.75439 Here, R is interpreting the dataframe penguins_numeric as a list, iterating through each column as an element of a list: 6.3.3.2 To do this with a for loop, we think about: What variable we need to loop through: the elements of penguins_numeric as a list. Do we need to store the outcome of this loop in a data structure? Yes, a numeric vector. At each iteration, what are we doing? Compute the mean of an element of penguins_numeric. result = rep(NA, ncol(penguins_numeric)) for(i in seq_along(penguins_numeric)) { result[i] = mean(penguins_numeric[[i]], na.rm = TRUE) } result ## [1] 43.92193 17.15117 200.91520 4201.75439 6.4 Exercises You can find exercises and solutions on Posit Cloud, or on GitHub. "],["about-the-authors.html", "About the Authors", " About the Authors These credits are based on our course contributors table guidelines. Credits Names Pedagogy Lead Content Instructor(s) Chris Lo Lecturer Chris Lo Content Author(s) (include chapter name/link in parentheses if only for specific chapters) - make new line if more than one chapter involved If any other authors besides lead instructor Content Contributor(s) (include section name/link in parentheses) - make new line if more than one section involved Wrote less than a chapter Content Editor(s)/Reviewer(s) Checked your content Content Director(s) Helped guide the content direction Content Consultants (include chapter name/link in parentheses or word “General”) - make new line if more than one chapter involved Gave high level advice on content Acknowledgments Gave small assistance to content but not to the level of consulting Production Content Publisher(s) Helped with publishing platform Content Publishing Reviewer(s) Reviewed overall content and aesthetics on publishing platform Technical Course Publishing Engineer(s) Helped with the code for the technical aspects related to the specific course generation Template Publishing Engineers Candace Savonen, Carrie Wright, Ava Hoffman Publishing Maintenance Engineer Candace Savonen Technical Publishing Stylists Carrie Wright, Ava Hoffman, Candace Savonen Package Developers (ottrpal) Candace Savonen, John Muschelli, Carrie Wright Art and Design Illustrator(s) Created graphics for the course Figure Artist(s) Created figures/plots for course Videographer(s) Filmed videos Videography Editor(s) Edited film Audiographer(s) Recorded audio Audiography Editor(s) Edited audio recordings Funding Funder(s) Institution/individual who funded course including grant number Funding Staff Staff members who help with funding ## ─ Session info ─────────────────────────────────────────────────────────────── ## setting value ## version R version 4.0.2 (2020-06-22) ## os Ubuntu 20.04.5 LTS ## system x86_64, linux-gnu ## ui X11 ## language (EN) ## collate en_US.UTF-8 ## ctype en_US.UTF-8 ## tz Etc/UTC ## date 2024-05-22 ## ## ─ Packages ─────────────────────────────────────────────────────────────────── ## package * version date lib source ## askpass 1.1 2019-01-13 [1] RSPM (R 4.0.3) ## assertthat 0.2.1 2019-03-21 [1] RSPM (R 4.0.5) ## bookdown 0.24 2024-03-13 [1] Github (rstudio/bookdown@88bc4ea) ## bslib 0.6.1 2023-11-28 [1] CRAN (R 4.0.2) ## cachem 1.0.8 2023-05-01 [1] CRAN (R 4.0.2) ## callr 3.5.0 2020-10-08 [1] RSPM (R 4.0.2) ## cli 3.6.2 2023-12-11 [1] CRAN (R 4.0.2) ## crayon 1.3.4 2017-09-16 [1] RSPM (R 4.0.0) ## desc 1.2.0 2018-05-01 [1] RSPM (R 4.0.3) ## devtools 2.3.2 2020-09-18 [1] RSPM (R 4.0.3) ## digest 0.6.25 2020-02-23 [1] RSPM (R 4.0.0) ## ellipsis 0.3.1 2020-05-15 [1] RSPM (R 4.0.3) ## evaluate 0.23 2023-11-01 [1] CRAN (R 4.0.2) ## fansi 0.4.1 2020-01-08 [1] RSPM (R 4.0.0) ## fastmap 1.1.1 2023-02-24 [1] CRAN (R 4.0.2) ## fs 1.5.0 2020-07-31 [1] RSPM (R 4.0.3) ## glue 1.4.2 2020-08-27 [1] RSPM (R 4.0.5) ## hms 0.5.3 2020-01-08 [1] RSPM (R 4.0.0) ## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.0.2) ## httr 1.4.2 2020-07-20 [1] RSPM (R 4.0.3) ## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.0.2) ## jsonlite 1.7.1 2020-09-07 [1] RSPM (R 4.0.2) ## knitr 1.33 2024-03-13 [1] Github (yihui/knitr@a1052d1) ## lifecycle 1.0.4 2023-11-07 [1] CRAN (R 4.0.2) ## magrittr 2.0.3 2022-03-30 [1] CRAN (R 4.0.2) ## memoise 2.0.1 2021-11-26 [1] CRAN (R 4.0.2) ## openssl 1.4.3 2020-09-18 [1] RSPM (R 4.0.3) ## ottrpal 1.2.1 2024-03-13 [1] Github (jhudsl/ottrpal@48e8c44) ## pillar 1.9.0 2023-03-22 [1] CRAN (R 4.0.2) ## pkgbuild 1.1.0 2020-07-13 [1] RSPM (R 4.0.2) ## pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.0.3) ## pkgload 1.1.0 2020-05-29 [1] RSPM (R 4.0.3) ## prettyunits 1.1.1 2020-01-24 [1] RSPM (R 4.0.3) ## processx 3.4.4 2020-09-03 [1] RSPM (R 4.0.2) ## ps 1.4.0 2020-10-07 [1] RSPM (R 4.0.2) ## R6 2.4.1 2019-11-12 [1] RSPM (R 4.0.0) ## readr 1.4.0 2020-10-05 [1] RSPM (R 4.0.2) ## remotes 2.2.0 2020-07-21 [1] RSPM (R 4.0.3) ## rlang 1.1.3 2024-01-10 [1] CRAN (R 4.0.2) ## rmarkdown 2.10 2024-03-13 [1] Github (rstudio/rmarkdown@02d3c25) ## rprojroot 2.0.4 2023-11-05 [1] CRAN (R 4.0.2) ## sass 0.4.8 2023-12-06 [1] CRAN (R 4.0.2) ## sessioninfo 1.1.1 2018-11-05 [1] RSPM (R 4.0.3) ## stringi 1.5.3 2020-09-09 [1] RSPM (R 4.0.3) ## stringr 1.4.0 2019-02-10 [1] RSPM (R 4.0.3) ## testthat 3.0.1 2024-03-13 [1] Github (R-lib/testthat@e99155a) ## tibble 3.2.1 2023-03-20 [1] CRAN (R 4.0.2) ## usethis 1.6.3 2020-09-17 [1] RSPM (R 4.0.2) ## utf8 1.1.4 2018-05-24 [1] RSPM (R 4.0.3) ## vctrs 0.6.5 2023-12-01 [1] CRAN (R 4.0.2) ## withr 2.3.0 2020-09-22 [1] RSPM (R 4.0.2) ## xfun 0.26 2024-03-13 [1] Github (yihui/xfun@74c2a66) ## xml2 1.3.2 2020-04-23 [1] RSPM (R 4.0.3) ## yaml 2.2.1 2020-02-01 [1] RSPM (R 4.0.3) ## ## [1] /usr/local/lib/R/site-library ## [2] /usr/local/lib/R/library "],["references.html", "Chapter 7 References", " Chapter 7 References "],["404.html", "Page not found", " Page not found The page you requested cannot be found (perhaps it was moved or renamed). You may want to try searching to find the page's new location, or use the table of contents to find the page you are looking for. "]]
diff --git a/docs/no_toc/writing-your-first-function.html b/docs/no_toc/writing-your-first-function.html
index f194cee..f8fa0f7 100644
--- a/docs/no_toc/writing-your-first-function.html
+++ b/docs/no_toc/writing-your-first-function.html
@@ -4,11 +4,11 @@
- Chapter 8 Writing your first function | Intermediate R, Season 3
-
+ Chapter 5 Writing your first function | Intermediate R, Season 3
+
-
+
@@ -16,7 +16,7 @@
-
+
@@ -29,8 +29,8 @@
-
-
+
+
@@ -138,6 +138,7 @@
Code that has a well-defined set of inputs and outputs make a good function.
A function should do only one, well-defined task.
-
-
8.0.1 Anatomy of a function definition
+
+
5.1 Anatomy of a function definition
Function definition consists of assigning a function name with a “function” statement that has a comma-separated list of named function arguments, and a return expression. The function name is stored as a variable in the global environment.
In order to use the function, one defines or import it, then one calls it.
Example:
@@ -276,13 +264,13 @@
8.0.1 Anatomy of a function defin
With function definitions, not all code runs from top to bottom. The first four lines defines the function, but the function is never run. It is called on line 5, and the lines within the function are executed.
When the function is called in line 5, the variables for the arguments are reassigned to function arguments to be used within the function and helps with the modular form. We need to introduce the concept of local and global environments to distinguish variables used only for a function from variables used for the entire program.
-
-
8.0.2 Local and global environments
+
+
5.2 Local and global environments
{ } represents variable scoping: within each { }, if variables are defined, they are stored in a local environment, and is only accessible within { }. All function arguments are stored in the local environment. The overall environment of the program is called the global environment and can be also accessed within { }.
The reason of having some of this “privacy” in the local environment is to make functions modular - they are independent little tools that should not interact with the rest of the global environment. Imagine someone writing a tool that they want to give someone else to use, but the tool depends on your environment, vice versa.
-
-
8.0.3 A step-by-step example
+
+
5.3 A step-by-step example
Using the addFunction function, let’s see step-by-step how the R interpreter understands our code:
@@ -301,8 +289,8 @@
8.0.3 A step-by-step example
We run the second line of code in the function body to return a value. The return value from the function is assigned to the variable z in the global environment. All local variables for the function are erased now that the function call is over.
-
-
8.0.4 Function arguments create modularity
+
+
5.4 Function arguments create modularity
First time writers of functions might ask: why are variables we use for the arguments of a function reassigned for function arguments in the local environment? Here is an example when that process is skipped - what are the consequences?
x = 3
y = 4
@@ -332,34 +320,42 @@
8.0.4 Function arguments create m
The function did not work as expected because we used hard-coded variables from the global environment and not function argument variables unique to the function use!
-
-
8.0.5 Exercises
+
+
5.5 Examples
Create a function, called add_and_raise_power in which the function takes in 3 numeric arguments. The function computes the following: the first two arguments are added together and raised to a power determined by the 3rd argument. The function returns the resulting value. Here is a use case: add_and_raise_power(1, 2, 3) = 27 because the function will return this expression: (1 + 2) ^ 3. Another use case: add_and_raise_power(3, 1, 2) = 16 because of the expression (3 + 1) ^ 2. Confirm with that these use cases work. Can this function used for numeric vectors?
-
add_and_raise_power =function(x, y, z) {
- result = (x + y)^z
-return(result)
-}
-add_and_raise_power(1, 2, 3)
+
add_and_raise_power =function(x, y, z) {
+ result = (x + y)^z
+return(result)
+}
+add_and_raise_power(1, 2, 3)
## [1] 27
Create a function, called my_dim in which the function takes in one argument: a dataframe. The function returns the following: a length-2 numeric vector in which the first element is the number of rows in the dataframe, and the second element is the number of columns in the dataframe. Your result should be identical as the dim function. How can you leverage existing functions such as nrow and ncol? Use case: my_dim(penguins) = c(344, 8)
Create a function, called medicaid_eligible in which the function takes in one argument: a numeric vector called age. The function returns a numeric vector with the same length as age, in which elements are 0 for indicies that are less than 65 in age, and 1 for indicies 65 or higher in age. Use cases: medicaid_eligible(c(30, 70)) = c(0, 1)
-
medicaid_eligible =function(age) {
- result = age
- result[age <65] =0
- result[age >=65] =1
-return(result)
-}
-medicaid_eligible(c(30, 70))
+
Create a function, called num_na in which the function takes in any vector, and then return a single numeric value. This numeric value is the number of NAs in the vector. Use cases: num_na(c(NA, 2, 3, 4, NA, 5)) = 2 and num_na(c(2, 3, 4, 5)) = 0. Hint 1: Use is.na() function. Hint 2: Given a logical vector, you can count the number of TRUE values by using sum(), such as sum(c(TRUE, TRUE, FALSE)) = 2.
Create a function, called medicaid_eligible in which the function takes in one argument: a numeric vector called age. The function returns a numeric vector with the same length as age, in which elements are 0 for indicies that are less than 65 in age, and 1 for indicies 65 or higher in age. (Hint: This is a data recoding problem!) Use cases: medicaid_eligible(c(30, 70)) = c(0, 1)
+
medicaid_eligible =function(age) {
+ result = age
+ result[age <65] =0
+ result[age >=65] =1
+return(result)
+}
+medicaid_eligible(c(30, 70))