Empress is a fast and scalable phylogenetic tree viewer.
To install the current development version, we recommend creating a new conda environment:
conda create -n empress python=3.6 numpy scipy pandas cython
conda activate empress
conda install -c bioconda scikit-bio biom-format
pip install git+https://github.com/biocore/empress.gitBefore following these instructions, make sure your QIIME 2 conda environment is activated (a version of at least 2019.1 is required). Then, run the following commands:
pip install git+https://github.com/biocore/empress.git
qiime dev refresh-cache
qiime empressIf you see information about Empress' QIIME 2 plugin, the installation was successful!
Empress can visualize Phylogeny[Rooted] QIIME 2 artifacts.
We're going to use data from the QIIME 2 Moving Pictures Tutorial:
rooted-tree.qzaview | downloadtable.qzaview | downloadtaxonomy.qzaview | downloadsample_metadata.tsvdownload
Once you've got the above files downloaded into a single directory, run the following command:
qiime empress plot \
--i-tree rooted-tree.qza \
--i-feature-table table.qza \
--m-sample-metadata-file sample_metadata.tsv \
--m-feature-metadata-file taxonomy.qza \
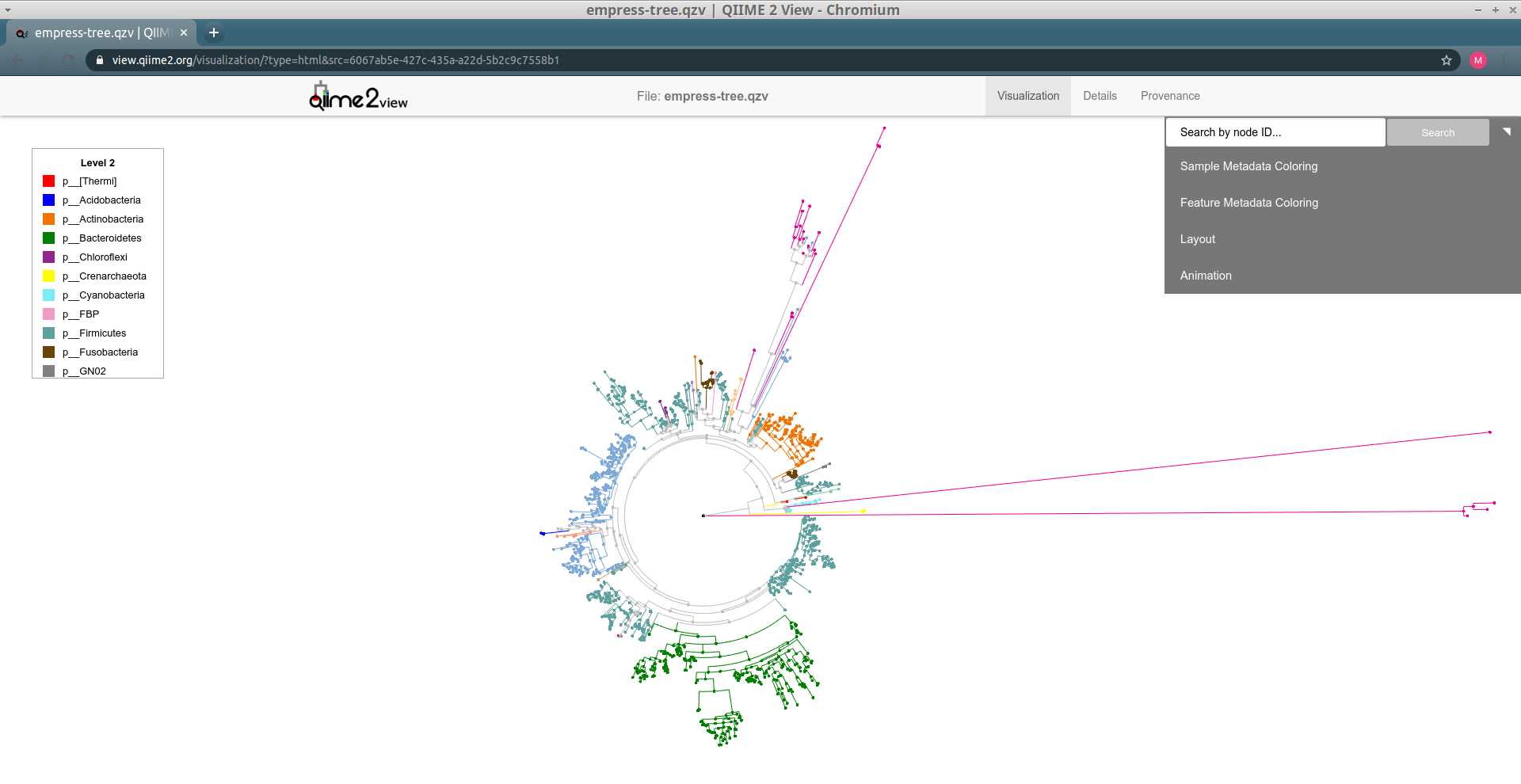
--o-visualization empress-tree.qzvThis QIIME 2 visualization can be viewed either using qiime tools view or by
uploading it to view.qiime2.org.
Please see the tests/README.md file for instructions on how to run Empress' tests.
This work is supported by IBM Research AI through the AI Horizons Network. For more information visit the IBM AI Horizons Network website.
Empress' JavaScript code is distributed with the source code of various
third-party dependencies (in the empress/support_files/vendor/ directory).
Please see
DEPENDENCY_LICENSES.md
for copies of these dependencies' licenses.